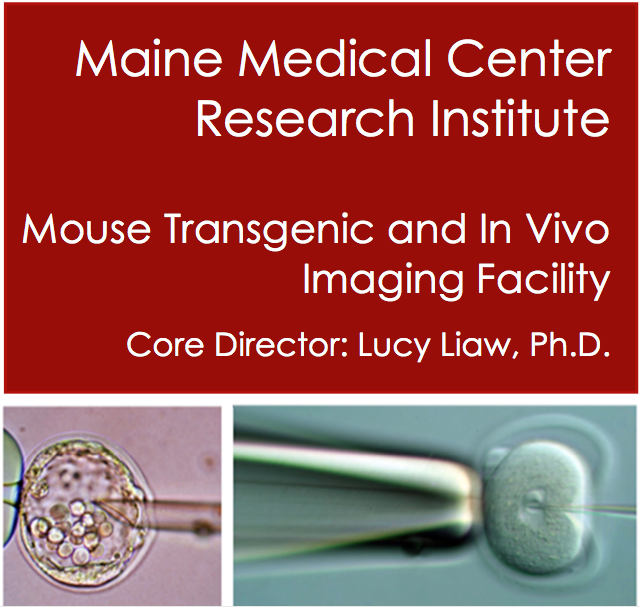
The Maine Medical Center Research Institute (MMCRI) has partnered with Tufts to provide a professional core facility that has over 15 years of experience providing high quality services for the generation of mouse transgenic strains including the use of CRISPR/Cas, cryopreservation of mouse germ cells, and imaging, including MRI and microCT. Mice are generated in a full barrier, AAALAC-accredited animal facility in a transgenic production room that facilitates direct importation of mice into the Tufts barrier facility. Contact us to discuss your mouse and imaging projects.
Mouse Transgenesis

We provide microinjection to generate your mouse models. Services include microinjection of fertilized oocytes with traditional DNA transgenes, or microinjection of CRISPR/Cas. ES cell injection is also performed. Contact us to design your CRISPR mouse project – cost depends on type of modification, strain of mouse, and days of injection. We have a high surveillance production room that will allow importation of mice direct into some barrier facilities.
Contact – Lucy Liaw, Ph.D., liawl@mmc.org
microCT
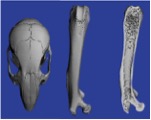
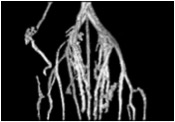
We house a Scanco high speed in vivo microCT scanner X-ray system. Our microCT facility has extensive experience in bone imaging and quantification, and can work on other projects where tissues are provided, i.e., vascular imaging of samples perfused with microfil. We provide quantification and any 3D images of the samples as required. Contact us to get a project quote. Pricing is based on hours of scanning and analysis time.
Contact – Lucy Liaw, Ph.D. liawl@mmc.org
Small Animal MRI
Our MRI facility houses a Bruker Pharmascan 7T, 300 MHz imager with 100 μm resolution. Services include anatomical imaging of most organs, angiography, proton spectroscopy and localized spectroscopy, and cardiac imaging, including diastolic and systolic dimensions of the ventricle. We can house “clean” animals at our facility for studies requiring longitudinal imaging. Contact us for mor information.
Contact – Ilka Pinz, Ph.D. pinzi@mmc.org