Featured Publications
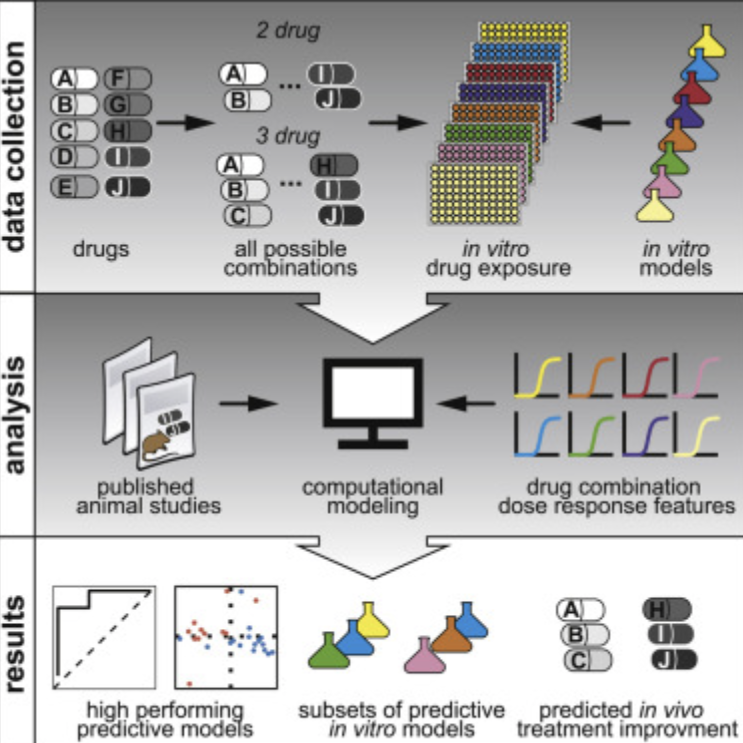
Jonah Larkins-Ford, Talia Greenstein, Nhi Van, Yonatan N. Degefu, Michaela C. Olson, Artem Sokolov, Bree B. Aldridge. “Systematic measurement of combination-drug landscapes to predict in vivo treatment outcomes for tuberculosis.” Cell Systems. August 31, 2021. [Source]
Trever C. Smith, Krista M. Pullen, Michaela C. Olson, Morgan E. McNellis, Ian Richardson, Sophia Hu, Jonah Larkins-Ford, Xin Wang, Joel S. Freundlich, D. Michael Ando, Bree B. Aldridge. “Morphological profiling of tubercle bacilli identifies drug pathways of action.” Proceedings of the National Academy of Sciences. July 2020, 202002738. DOI: 10.1073/pnas.2002738117 [Source]
Logsdon, M., Ho, P., Papavinasasundaram, K., Richardson, K., Cokol, M., Sassetti, C., Amir, A., Aldridge, B.B. “A parallel adder coordinates mycobacterial cell cycle progression and cell size homeostasis in the context of asymmetric growth and organization.” Current Biology. 6 November 2017; 27:1-8. [Source]
Pre-Print
Mycobacterium tuberculosis grows linearly at the single-cell level with larger variability than model organisms. Eun Seon Chung, Prathitha Kar, Maliwan Kamkaew, Ariel Amir, Bree B Aldridge. link
Massively parallel combination screen reveals small molecule sensitization of antibiotic-resistant Gram-negative ESKAPE pathogens. Megan W Tse, Meilin Zhu, Benjamin Peters, Efrat Hamami, Julie Chen, Kathleen P Davis, Samuel Nitz, Juliane Weller, Thulasi Warrier, Diana K Hunt, Yoelkys Morales, Tomohiko Kawate, Jeffrey L Gaulin, Jon H Come, Juan Hernandez-Bird, Wenwen Huo, Isabelle Neisewander, Laura L Kiessling, Deborah T Hung, Joan Mecsas, Bree B Aldridge, Ralph R Isberg, Paul C Blainey. bioRxiv 2024.03.26.586803; link.
Unique growth and morphology properties of Clade 5 Clostridioides difficile strains revealed by single-cell time-lapse microscopyJohn W. Ribis, César A. Nieto-Acuña, Qiwen Dong, Nicholas V. DiBenedetto, Anchal Mehra, Irene Nagawa, Imane El Meouche, Bree B. Aldridge, Mary J. Dunlop, Rita Tamayo, Abyudhai Singh, Aimee Shen. bioRxiv 2024.02.13.580212; link
Publications
Davis KP, Morales Y, Ende RJ, Peters R, McCabe AL, Mecsas J, Aldridge BB. 2024. Critical role of growth medium for detecting drug interactions in Gram-negative bacteria that model in vivo responses. mBio 15:e00159-24. link
Semi-automated colony-forming unit counting for biosafety level 3 laboratories. Soorya Janakiraman, Sophie Engels, Pariksheet Nanda, Maral Budak, Talia Greenstein, Mariana Pereira Moraes, Bree B. Aldridge, Denise E. Kirschner. STAR Protocols (2023).
Novel synergies and isolate specificities in the drug interactions landscape of Mycobacterium abscessus. Nhi Van, Yonatan N Degefu, Pathricia A Leus, Jonah Larkins-Ford, Jacob Klickstein, Florian P Maurer, David Stone, Husain Poonawala, Cheleste M Thorpe, Trever C Smith II, Bree B Aldridge. Antimicrob Agents Chemother. 2023 Jun 6;e0009023. doi: 10.1128/aac.00090-23. link
Confusion. (Can we do better when it comes to the “other-race effect”?) K Heran Darwin, Bree Aldridge, Jessica Seeliger, Aimee Shen, Sarah Stanley. EMBO Rep. 2023 Apr 5;24(4):e57041. link
How can systems approaches help us understand and treat infectious disease? Anna Kuchina, Jason Yang, Bree B. Aldridge, Kevin A. Janes, Naeha Subramanian, Nevan J. Krogan, Mehdi Bouhaddou, Shirit Einav, Jason Papin, Ronald N. Germain. Cell Systems (2022). link
Advances in the design of combination therapies for the treatment of tuberculosis. Jonah Larkins-Ford and Bree B. Aldridge. Expert Opinion on Drug Discovery (2022). link
Tools to develop antibiotic combinations that target drug tolerance in Mycobacterium tuberculosis. Talia Greenstein and Bree B. Aldridge. Frontiers in Cellular and Infection Microbiology (2022). link
Tuberculosis treatment failure associated with evolution of antibiotic resilience. Qingyun Liu, Junhao Zhu, Charles L. Dulberger, Sydney Stanley, Sean Wilson, Eun Seon Chung, Xin Wang, Peter Culviner, Yue J. Liu, Nathan D. Hicks, Gregory H. Babunovic, Samantha R. Giffen, Bree B. Aldridge, Ethan C. Garner, Eric J. Rubin, Michael C. Chao, Sarah M. Fortune. Science. 2022 Dec 9;378(6624):1111-1118. Epub 2022 Dec 8. link
Design principles to assemble drug combinations for effective tuberculosis therapy using interpretable pairwise drug response measurements. Jonah Larkins-Ford, Yonatan N. Degefu, Nhi Van, Artem Sokolov, Bree B. Aldridge. Cell Reports Medicine (2022). link
- Preview: Predicting in vivo activity of combination therapies from in vitro drug pairs in diverse environments
- Press: Tufts Scientists Use Artificial Intelligence to Improve Tuberculosis Treatments
C25-modified rifamycin derivatives with improved activity against Mycobacterium abscessus. Laura Paulowski, Katherine S H Beckham, Matt D Johansen, Laura Berneking, Nhi Van, Yonatan Degefu, Sonja Staack, Flor Vasquez Sotomayor, Lucia Asar, Holger Rohde, Bree B Aldridge, Martin Aepfelbacher, Annabel Parret, Matthias Wilmanns, Laurent Kremer, Keith Combrink, Florian P Maurer. PNAS Nexus, Volume 1, Issue 4 (2022). link
Localization of EccA3 at the growing pole in Mycobacterium smegmatis. Nastassja L. Kriel, Mae Newton-Foot, Owen T. Bennion, Bree B. Aldridge, Carolina Mehaffy, John T. Belisle, Gerhard Walzl, Robin M. Warren, Samantha L. Sampson & Nico C. Gey van Pittius. BMC Microbiology volume 22, Article number: 140 (2022). link
Eun Seon (Christin) Chung, William C. Johnson, Bree B. Aldridge.
Types and functions of heterogeneity in mycobacteria. Nature Reviews Microbiology (2022). link
Kathleen Davis, Talia Greenstein, Roberto Viau Colindres, Bree B Aldridge.
Leveraging laboratory and clinical studies to design effective antibiotic combination therapy, Current Opinion in Microbiology (2021). Volume 64: 68-75. link
Li et al., Identification of cell wall synthesis inhibitors active against Mycobacterium tuberculosis by competitive activity-based protein profiling, Cell Chemical Biology (2021), DOI.
Jonah Larkins-Ford, Talia Greenstein, Nhi Van, Yonatan N. Degefu, Michaela C. Olson, Artem Sokolov, Bree B. Aldridge. Systematic measurement of combination-drug landscapes to predict in vivo treatment outcomes for tuberculosis. Cell Systems. August 31, 2021. DOI.
Egbelowo O, Sarathy JP, Gausi K, Zimmerman MD, Wang H, Wijnant GJ, Kaya F, Gengenbacher M, Van N, Degefu Y, Nacy C, Aldridge BB, Carter CL, Denti P, Dartois V. Pharmacokinetics and target attainment of SQ109 in plasma and human-like tuberculosis lesions in rabbits. Antimicrob Agents Chemother. 2021 Jul 6:AAC0002421. Epub ahead of print. PMID: 34228540. DOI.
Van N., Degefu Y.N., Aldridge B.B. (2021) Efficient Measurement of Drug Interactions with DiaMOND (Diagonal Measurement of N-Way Drug Interactions). In: Parish T., Kumar A. (eds) Mycobacteria Protocols. Methods in Molecular Biology, vol 2314. Humana, New York, NY. DOI
Aldridge, B.B., Barros-Aguirre, D., Barry, C.E. et al. The Tuberculosis Drug Accelerator at year 10: what have we learned?. Nat Med (2021). DOI
Trever C. Smith, Krista M. Pullen, Michaela C. Olson, Morgan E. McNellis, Ian Richardson, Sophia Hu, Jonah Larkins-Ford, Xin Wang, Joel S. Freundlich, D. Michael Ando, Bree B. Aldridge. Morphological profiling of tubercle bacilli identifies drug pathways of action. Proceedings of the National Academy of Sciences. July 2020, 202002738. DOI: 10.1073/pnas.2002738117 link
Ardeshir Goliaei, Haley A Woods, Adriana E Tron, Matthew A Belmonte, J Paul Secrist, Douglas Ferguson, Lisa Drew, Adrian J Fretland, Bree B Aldridge, Francis D Gibbons. Multiscale Model Identifies Improved Schedule for Treatment of Acute Myeloid Leukemia In Vitro With the Mcl‐1 Inhibitor AZD5991. CPT: pharmacometrics & systems pharmacology. October 2020. DOI: https://doi.org/10.1002/psp4.12552
Shuyi Ma, Suraj Jaipalli, Jonah Larkins-Ford, Jenny Lohmiller, Bree B. Aldridge, David R. Sherman, Sriram Chandrasekaran. Transcriptomic Signatures Predict Regulators of Drug Synergy and Clinical Regimen Efficacy against Tuberculosis. mBio. Nov 2019, 10 (6) e02627-19 link
Manu De Rycker, David Horn, Bree Aldridge, et al., Setting Our Sights on Infectious Diseases. ACS Infectious Diseases. 2019. link
Smith TC, Aldridge BB. Targeting drugs for tuberculosis. 28
June 2019. Science. Vol. 364, Issue 6447, pp. 1234. free full text
Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019. link
Katzir, I., Cokol, M., Aldridge, B.B.*, Alon, U.* “Prediction of ultra-high-order antibiotic combinations based on pairwise interactions.” (2019) PLoS Computational Biology 15(1):e1006774. link
Boot, M., Commandeur, S., Subudhi, A.K., Bahira, M., Smith II, T.C., Abdallah, A.M., van Gemert, M., Lelièvre, J., Ballell, L., Aldridge, B.B, Pain, A., Speer, A., and Bitter, W. “Accelerating early anti-tuberculosis drug discovery by creating mycobacterial indicator strains that predict mode of action.” Antimicrobial Agents and Chemotherapy (in press). link
Logsdon, M.M. and Aldridge, B.B. “Stable Regulation of Cell Cycle Events in Mycobacteria: Insights from Inherently Heterogeneous Bacterial Populations.” (2018) Frontiers in Microbiology, 9:514. link
Hayashi, J.M., Richardson, K., Melzer, E.S., Sandler, S.J., Aldridge, B.B., Siegrist, M.S., Morita, Y.S. (2018). “Stress-Induced Reorganization of the Mycobacterial Membrane Domain.” mBio, 9(1). link
Sundarakrishnan, A., Chen, Y., Black, L. D., Aldridge, B. B. , Kaplan, D.L. (2018). “Engineered cell and tissue models of pulmonary fibrosis” Advanced Drug Delivery Reviews, in press. link
Mann, K. M., Huang, D. L., Hooppaw, A. J., Logsdon, M. M., Richardson, K., Lee, H. J., Kimmey, J. M., Aldridge, B. B. , Stallings, C. (2017). “Rv0004 is a new essential member of the mycobacterial DNA replication machinery” PLoS Genetics, 13(11), e1007115. link
Srinivasan Vijay, Dao N. Vinh, Hoang T. Hai, Vu T. N. Ha, Vu T. M. Dung, Tran D. Dinh, Hoang N. Nhung, Trinh T. B. Tram, Bree B. Aldridge, Nguyen T. Hanh, Do D. A. Thu, Nguyen H. Phu, Guy E. Thwaites, Nguyen T. T. Thuong. “Influence of Stress and Antibiotic Resistance on Cell-Length Distribution in Mycobacterium tuberculosis Clinical Isolates” Frontiers in Microbiology. 21 November 2017. link
Logsdon, M., Ho, P., Papavinasasundaram, K., Richardson, K., Cokol, M., Sassetti, C., Amir, A., Aldridge, B.B. “A parallel adder coordinates mycobacterial cell cycle progression and cell size homeostasis in the context of asymmetric growth and organization” Current Biology. 6 November 2017; 27:1-8. link
Cokol, M., Kuru, N., Bicak, E., Larkins-Ford, J., Aldridge, B.B. “Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis” Science Advances. 2017 October 11: E1701881. link
Richardson, K., Bennion, O.T., Tan, S., Hoang, A.N., Cokol, M., Aldridge, B.B. “Temporal and intrinsic factors of rifampicin tolerance in mycobacteria” Proc Natl Acad Sci U S A. 2016 June 29. link
Hayashi, J.M., Luo, C.Y., Mayfield, J.A., Hsu T., Fukuda, T., Walfield, A.L., Giffen, S.R., Leszyk, J.D., Baer, C.E., Bennion, O.T., Madduri, A., Shaffer, S.A., Aldridge, B.B., Sassetti, C.M., Sandler, S.J., Kinoshita, T., Moody, D.B., Morita, Y.S. “Spatially distinct and metabolically active membrane domain in mycobacteria.” Proc Natl Acad Sci U S A. 2016 May 10;113(19):5400-5. link
Aldridge B.B., Keren I., Fortune S.M. “The Spectrum of Drug Susceptibility in Mycobacteria. “Microbiol Spectr. 2014 Oct;2(5). link
Aldridge B.B., Rhee K.Y. “Microbial metabolomics: innovation, application, insight.” Curr Opin Microbiol. 2014 Jun;19:90-6. link
Chao, M.C., Kieser, K.J., Minami, S, Mavrici, D., Aldridge, B.B., Fortune, S.M., Alber, T., Rubin, E.J. “Protein Complexes and Proteolytic Activation of the Cell Wall Hydrolase RipA Regulate Septal Resolution in Mycobacteria.” PLoS Pathogens, 2013, 9(2): e1003197. link
Wirth, S.E., Krywy, J.A., Aldridge, B.B., Fortune, S.M., Fernandez-Suarez, M., Gray, T.A., Derbyshire, K.M., “Polar assembly and scaffolding proteins of the virulence-associated ESX-1 secretory apparatus in mycobacteria.” Molecular Microbiology, 2012, 83(3):654-664. PMCID: PMC3277861 link
Aldridge, B.B., Fernandez-Suarez, M., Heller, D., Ambravaneswaran, V., Irimia, D., Toner, M., Fortune, S.M., “Asymmetry and aging in mycobacterial cells lead to variable growth and antibiotic susceptibility.” Science, 2012, 335 (6064): 100-104. PMCID: PMC3397429 link
Aldridge, B.B., Gaudet, S., Lauffenburger, D.A., Sorger, P.K., “Lyapunov exponents and phase diagrams reveal multi-factorial control over TRAIL-induced apoptosis.” Molecular Systems Biology, 2011, 7:553. PMCID: PMC3261706 link
Aldridge, B.B., Saez-Rodriguez, J., Muhlich, J., Sorger, P.K., Lauffenburger, D.A., “Fuzzy logic analysis of kinase pathway crosstalk in TNF/EGF/Insulin-induced signaling.” PLoS Computational Biology, 2009, 5(4): e1000340. PMCID: PMC2663056 link
Albeck, J.G., Burke, J.M., Aldridge, B.B., Zhang, M.S., Lauffenburger, D.A., Sorger, P.K. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells.” Mol. Cell, 2008, 30(1):11-25. PMCID: PMC2858979 link
Aldridge, B.B., Burke, J., Lauffenburger, D., Sorger, P. Physicochemical Modelling of Cell Signaling Pathways. Nature Cell Biol, 2006, 8:1195-2003. link
Aldridge, B.B., Haller, G., Sorger, P., and Lauffenburger, D. Direct Lyapunov exponent analysis enables parametric study of transient signalling governing cell behaviour. Syst. Biol, 2006. 153 (6). link
Draviam VM, Shapiro I, Aldridge B., and Sorger PK. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 2006. 25 (12): 2814-27. PMCID: PMC1500857 link
