DNA Methylation Mechanism
The methylation of C5 on cytosine is catalyzed by DNA Methyltransferase (C5-MTases) (Fig. 1, 2)[14][15].
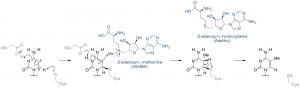
It requires a methyl group donating cofactor: S-adenosyl-L-methionine (AdoMet, SAM) (Fig. 3)[15]. The process of methylation consists of the following steps (Fig. 4)[11].

Fig 2. Cysteine residue at DNA Methyltransferase active site[15]
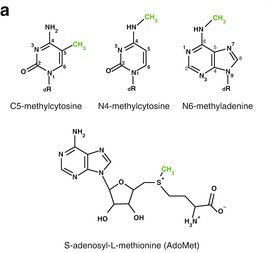
Fig. 3 C5 Position and AdoMet[15]
- Cysteine residue of the motif IV (PCQ) initiates nucleophilic attack on the C6 position of the cytosine ring, forming a covalent bond with the sixth carbon of the target cytosine base [12].
- Subsequently transfer of a methyl group from AdoMet to the C5 carbon of cytosine. The glutamate from the ENV motif stabilizes the base via the contact with the N4 exocyclic amino group. [13] AdoMet loses a methyl group and becomes S-Adenosyl-L-homocysteine (AdoHcy, SAH).
- Proton abstraction from the transient covalent intermediate by a catalyti general base on enzyme C5-MTases [13], eliminating the Cysteine side chain of the enzyme and restore the C5-6 double bond.
November 13, 2017 at 8:03 pm
It’s cool to see molecules from lectures outside of the class material! I think this page is pretty solid, one small suggestion, from a purely formatting perspective, would be to maybe group the figures together into a single block to prevent the need to scroll up and down when referencing figures.