PGC7: Protection Against Demethylation during Gametogenesis
Nakamura, Toshinobu, et al. “PGC7 Binds Histone H3K9me2 to Protect against Conversion of 5mC to 5hmC in Early Embryos.” Nature, vol. 486, no. 7403, 2012, pp. 415–9. [25]
The experiment on pre-diabetic F0 mice by Yanchang Wei, et al. suggested that some epigenetic marks on the F0 germ cells evade demethylation processes during gametogenesis and early embryogenesis, resulting in impaired glucose tolerance and insulin resistance in F1 and F2. This paper by Toshinobu Nakamura, et al. explored a specific mechanism for resistance of the methylated marks to reprogramming.
Methylation of the cytosine base results in 5-methylcytosine (5mC), and the oxidation from 5mC to 5-hydroxymethylcytosine (5 hmC) by TET enzymes leads to a passive pathway of demethylation. [10] In this experiment with early embryos, data indicated that the maternal factor PGC7 protected 5mC from demethylation and provided a mechanism for the prevention of TET-mediated oxidation.
BINDING STRENGTH OF PGC7 TO H3K9me2
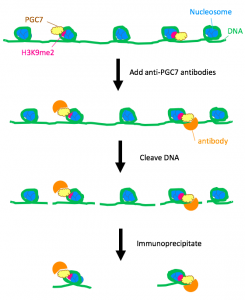
PGC7 is factor that is present in only oocytes. The first step was to determine the binding strength of PGC7 to histone H3K9me2 (histone 3, dimethylated at lysine 9) using mouse embryonic stem (ES) cells. A chromatin-immunoprecipitation (ChIP) assay in wild-type ES cells and PGC7-knockout ES cells using anti-PGC7 antibody revealed that the nucleosomes bound to PGC7 had H3K9me2.
Figure 1. ChIP assay of wild-type mouse ES cells. All wild-type ES cells express PGC7, and the results of immunoprecipitation indicated that PGC7 bound nucleosomes containing the histone H3K9me2.
Furthermore, in-vitro mixtures of PGC7 with different histones, as well as a competitive binding assay, demonstrated that H3K9me2 bound to PGC7 with the greatest strength. This finding suggested that in the cell, PGC7 directly bound to the H3K9me2 histones.
ROLE OF PGC7 DURING DNA DEMETHYLATION
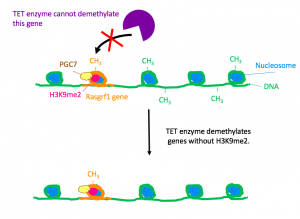
The next step was to determine the role of PGC7 and histone H3K9me2 in protection against DNA demethylation. The researchers studied the paternally imprinted genes H19 and Rasgrf1 in ES cells. Previously, researchers had known that DNA methylation at H19 and Rasgrf1 are usually conserved throughout embryogenesis. In this experiment, the researchers fertilized PGC7-muted oocytes with wild-type sperm and found that after embryogenesis, those genes lost methylation. Furthermore, ChIP analysis showed an abundance of H3K9me2 at those genes. These observations showed that PGC7 binds to loci attached H3K9me2, protecting the methylated genes from demethylation by TET enzymes.
Figure 2. Wild-type, PGC7-expressing cell. In the presence of PGC7, the TET enzyme cannot demethylate the Rasgrf1 gene.
Finally, the researchers investigated the impact of PGC7 on the binding between a TET enzyme and chromatin using a stepwise salt-extraction analysis on PGC7+ and PGC7- ES cells. In the wild-type, PGC7 weakened the binding between Tet3 and the chromatin. With this finding, the authors hypothesized that upon binding to H3K9me2, PGC7 induces changes in chromatin configuration. These changes prevent TET enzymes from attaching to DNA and demethylating imprinted genes.
CONCLUSION
The results of this study showed that PGC7 strongly binds to histone H3K9me2, and that this binding prevents TET enzymes from adhering to the chromatin. As previously described, experiments on F0 pre-diabetic mice by Yanchang Wei, et al. showed similarities in the methylomes of the F0 sperm and F1 pancreatic islets, indicating the inheritance of methylated marks. This form of epigenetic inheritance depends on the resistance of the methylated marks against reprogramming during gametogenesis and embryogenesis. The findings of Nakamura, et. al elucidated a possible mechanism that protects imprinted genes from demethylation, allowing epigenetic inheritance.
Leave a Reply