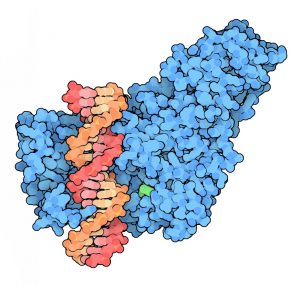
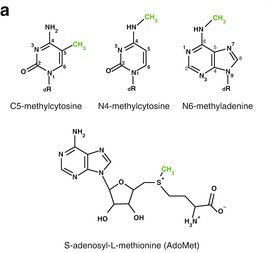
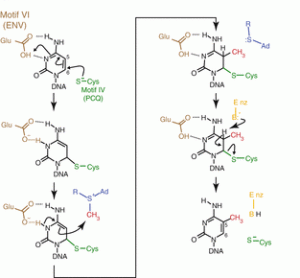
The methylation of C5 on cytosine is catalyzed by DNA Methyltransferase (C5-MTases) (Fig. 1, 2). It requires a methyl group donating cofactor: S-adenosyl-L-methionine (AdoMet, SAM) (Fig. 3). The process of methylation consists of the following steps (Fig. 4)
- Cysteine residue of the motif IV (PCQ) initiates nucleophilic attack on the C6 position of the cytosine ring, forming a covalent bond with the sixth carbon of the target cytosine base (Song et al. 2012)
- Subsequently transfer of a methyl group from AdoMet to the C5 carbon of cytosine. The glutamate from the ENV motif stabilizes the base via the contact with the N4 exocyclic amino group (Lukashevich, Olga V. et al. 2017). AdoMet loses a methyl group and becomes S-Adenosyl-L-homocysteine (AdoHcy, SAH).
- Proton abstraction from the transient covalent intermediate by an unknown general base on enzyme C5-MTases (Lukashevich, Olga V. et al. 2017), eliminating the Cysteine side chain of the enzyme and restore the C5-6 double bond.
Referance
- Lukashevich, Olga V. et al. “Conserved Motif VIII of Murine DNA Methyltransferase Dnmt3a Is Essential for Methylation Activity.” BMC Biochemistry 17 (2016): 7. PMC. Web. 8 Oct. 2017.
- Song, Jikui et al. “Structure-Based Mechanistic Insights into DNMT1-Mediated Maintenance DNA Methylation.” Science (New York, N.Y.) 335.6069 (2012): 709–712. PMC. Web. 8 Oct. 2017.
- Figure 1. https://blogs.biomedcentral.com/on-biology/2013/12/05/dna-methyltransferase-inhibitors-shifting-realities/
- Figure 2, 3, 4. Jeltsch, A. (2016) DNA methyltransferases: Role and function. SpringerCham.

Recent Comments