DNA Demethylation Pathway
DNA Demethylation
DNA demethylation is involved in many crucial biological process such as gene expression, gene silencing, and embryonic development. [8] This epigenetic modification pathway can result in transposable element silencing, genome imprinting, and X chromosome inactivation. [9] Active DNA demethylation occurs during specific time in early development. For example, gene expression marks are actively regulated during early embryonic stem cell development in order to progress in the transcriptional programme that leads to the development of specific cell type. [10]
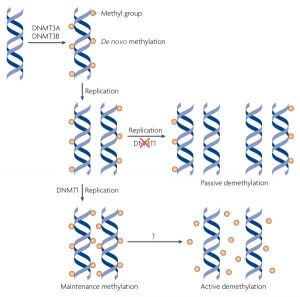
Passive Demethylation
DNA methyltransferas DNMT 1 recognizes 5-methylcytosine but not 5-hydroxymethylcytosine. Oxidation of 5-methylcytosine to 5-hydroxymethylcytosine by 5-carboxycytosine by ten-eleven translocation (TET) 5-methylcytosin deoxgenase enzyme or other oxidative agents provides a passive pathway for demethylation. The base-pairing chemistry for 5-Hydroxymethylcytosine is the same as cytosine and 5-methylcytosine, so it will still be replicated correctly to cytosine. [10]
Active Demethylation

Fig 2. Active DNA Demethylation [9]
November 13, 2017 at 6:16 pm
A bit of clarification might be needed for the passive demethylation section. Does oxidation of 5-methylcytosine to 5-hydroxymethylcytosine occur ‘spontaneously’ in the nucleus, or does some other non-ATP dependent molecule passively oxidize 5-methylcytosine?