MeDIP-Seq: A Molecular Method to Study Epigenetics
Methylated DNA Immunoprecipitation (MeDIP) isolates methylated DNA regions in the genome using affinity binding assay. MeDIP can be combined with next generation sequencing (MeDIP-seq) to locate the methylated DNA regions.
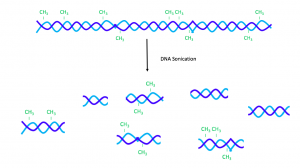
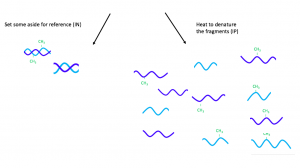
To begin MeDIP, the DNA sample is divided into 300 – 1000 base-pair fragments by sonication. [20] Then the fragments are divided into two groups — reference (IN) and immunoprecipitation (IP). While the IN fragments are set aside, the IP ones are heated to denaturation.
Figure 1. Sonication and denaturation
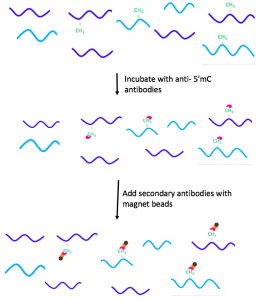
The IP sample is then incubated with anti-5’mC, which is a type of monoclonal antibody that binds methylated DNA. [20] As the next step, the IP sample is mixed with a secondary antibody, which is attached to magnetic beads. [20]The secondary antibody also has affinity to the anti-5’mC and binds it. [20]
Figure 2. Addition of antibodies
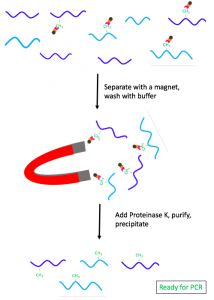
Once the antibody complex binds the targeted regions, the methylated DNA fragments are extracted using a magnet. The IP sample is washed with buffer to remove the non-methylated DNA and incubated with Proteinase K to degrade the antibodies. [20] The sample is purified using phenol:chloroform extraction and then precipitated in water. [20] Finally, the IN and IP samples are amplified using PCR and compared to identify the methylated regions of the DNA. [20]
Figure 3. Separation and purification of the methylated fragments.
The MeDIP procedure may be combined with next generation sequencing (MeDIP-seq). [21] The DNA sample is first divided by sonification, and the fragments are then prepared for library preparation through adapter ligation. [21] The aforementioned immunoprecipitation procedure is used to extract the methylated DNA regions, and adapter-mediated PCR is performed with the sample. [21] After DNA purification, next-generation sequencing is performed with the PCR product. [21]
Leave a Reply