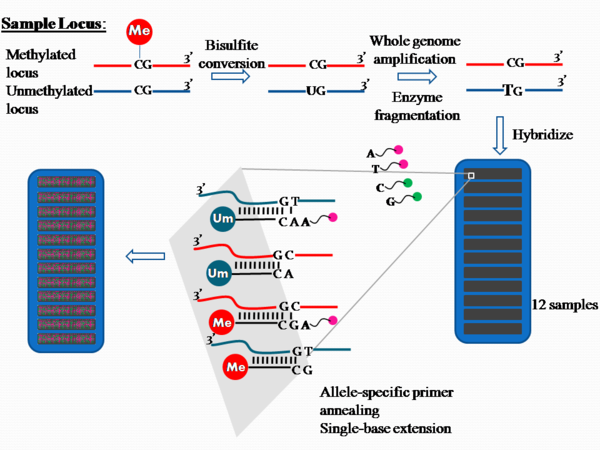
PCR amplification and fluorophore sequencing are common DNA sequencing techniques in the biomedical field. It is the bisulfite conversion step that sets the Illumina method apart and allows DNA sequences to be examined for methylation patterns. After this conversion step the product contains unconverted cytosine if they were previously methylated, but cytosine is converted to uracil if they were previously unmethylated (24).
Mehta et. al use this method in their paper studying Vietnam war veterans to determine the relative amount of methylation at specific gene loci.
- Bisulfite Conversion (Pre-Amplification): DNA is treated with sodium bisulfite using an Illumina specified bisulfite conversion kit. After the treatment, unmethylated cytosines convert to uracil, though methylated cytosines remain unchanged.
- BCD samples are denatured and neutralized.
- The denatured DNA is amplified by PCR and then fragmented by endpoint fragmentation
- Hybridize to BeadChip (Post-Amplification): The fragmented DNA samples are dispensed onto BeadChips. Each BeadChip is a 3-micron silica bead. Together these BeadChips can self assemble into a microarray with a uniform spacing of ~5.7 microns.
- The DNA samples are incubated to hybridize the samples onto the BeadChips. The amplified and fragmented DNA samples anneal to locus-specific 50mers during hybridization. Two bead types correspond to each CpG locus for the assay: one bead type corresponds to methylated (C), another bead type to unmethylated (T) state of the CpG site.
Each bead is covered with hundreds of thousands of copies of a specific oligonucleotide that act as the capture sequences in a given Illumina assay.
- The DNA samples are incubated to hybridize the samples onto the BeadChips. The amplified and fragmented DNA samples anneal to locus-specific 50mers during hybridization. Two bead types correspond to each CpG locus for the assay: one bead type corresponds to methylated (C), another bead type to unmethylated (T) state of the CpG site.
- Extend and Stain BeadChip (Post-Amplification): The chip undergoes extension and staining with anti-DNP-red and streptavidin-green in capillary flow-through chambers. Single-base extension of the oligos on the BeadChip, using the captured DNA as a template, incorporates detectable labels on the BeadChip and determines the methylation level of the query CpG sites.
- Image BeadChip (Post-Amplification): The Illumina HiScan scans the BeadChip, using a laser to excite the fluorophore of the single-base extension product on the beads. The scanner records high resolution images of the light emitted from the fluorophores.
- These images show the intensities of the methylated and unmethylated bead types, and the fluorescence intensity ratios between the two bead types are calculated.
- For a given individual at a given locus, a ratio value of 0 equals to non-methylation of the locus (homozygous unmethylated); a ratio of 1 equals to total methylation (homozygous methylated); and a value of 0.5 means that one copy is methylated and the other is not (i.e., heterozygosity), in the diploid human genome.

1) In what way does the bisulfite conversion set the Illumina method apart?
2) What are BeadChips?
3) Where is this fluorophore from?
4) What, specifically, does the light information collected by the HiScan mean?
Otherwise, good job!
Is there a reason why some steps are bolded and not others?
What does the last step in Figure 22 show?
It may be helpful to label Figure 22 with the numbers that are used to list the steps above.