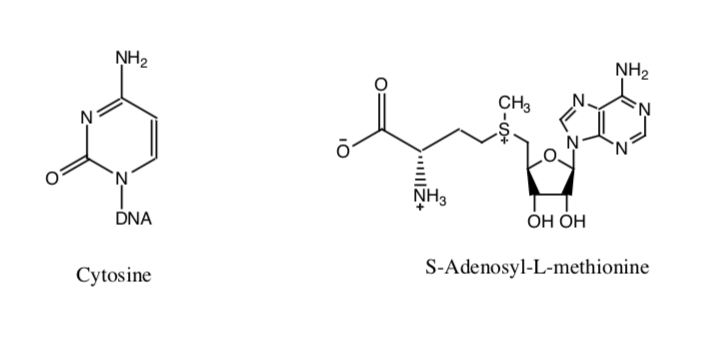
The DNA methylation reaction requires two vital substrates: cytosine and the cofactor S-Adenosyl-L-methionine, as shown above (Figure 16) (19). As noted in the DNA Methylation background page, DNMT uses two important residues to help stabilize the base in the active site (Cys-1226 and Glu-1266), as seen in Figure 17 (19).
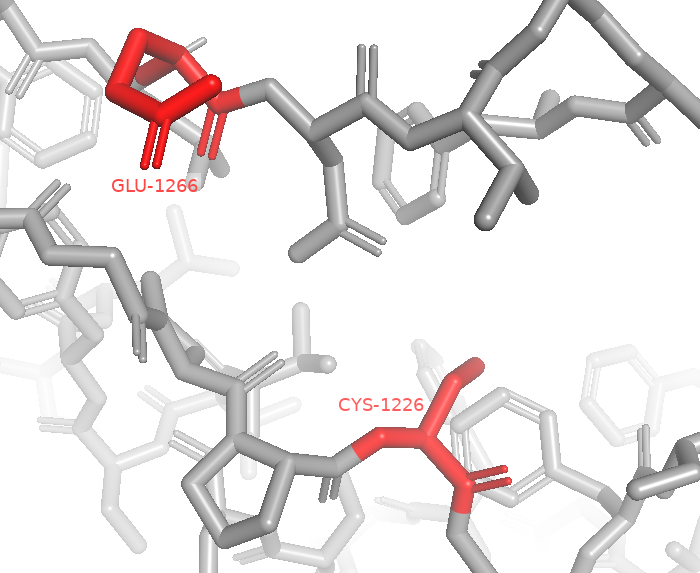
and proton transfers with the cofactors. (PDB-3PTA) (12).
Taking place in the active site of DNA methyltransferase, the methylation reaction is proposed to run as the following:
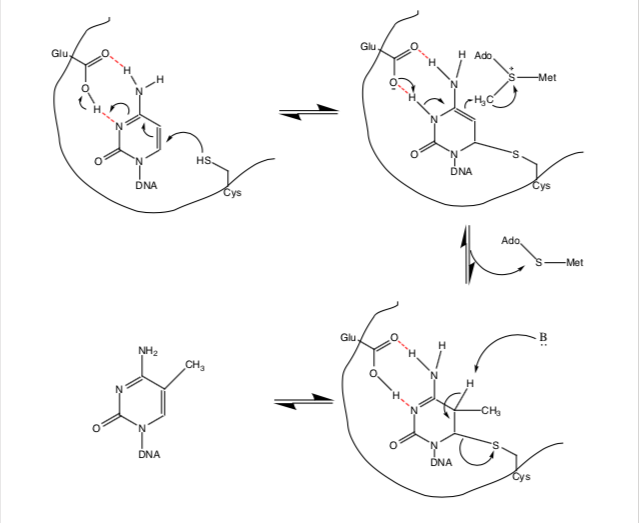
- Cytosine is stabilized by a Glu-1266 reside in the active site of DNMT, held close to a nucleophilic Cys-1226 residue.
- The Cys-1226 residue nucleophilically attacks the C6 on cytosine, causing double bonds in the ring to rotate, eventually leading to a proton transfer between the Glu-1266 residue and N3 on cytosine.
- The cofactor SAM enters the active site.
- The proton on N3 is then transferred back to the negatively-charged Glu-1266 residue, causing the double bond to reform between N3 and C4; this, in turn, causes alkylation by the SAM methyl group.
- An unknown basic residue then deprotonates the remaining proton on C5, reforming the double bond between C5 and C6.
- The DNA-protein complex then breaks down and aromaticity is restored; the Cys-1226 bond is broken, leading to the methylated cytosine, or 5-methylcytosine (19).

nicely done! I like that you use the pymol figures to show the enzyme as well as the mechanism figures. Very clear.
The reaction mechanism with the arrow pushing makes this biochemical pathway very easy to understand
This was very concise! Consider adding small numbers to Figure 19. to signify the appropriate steps.
Maybe considering briefly mentioning again how the two residues in DNMT play a role in the mechanism instead of just referring back to the previous page.
Also you accidentally wrote DMNT instead of DNMT in Figure 19.
Having the description of steps along with the visual arrow-pushing is very helpful!
Done