Technically, all cells of eukaryotes contain the same genetic information (DNA); Yet, all cells do not express the same genes. Thus, DNA Methylation (one of many epigenetic mechanisms) aids in the differential expression of genes depending on cell type.
The major type of DNA methylation occurs on cytosines in CpG islands, which are portions of DNA where cytosines are followed immediately by guanines (Figure 5). (9).
Before DNA methylation occurs, cytosines must be turned out of the DNA double helix so that they can be exposed. In some cases, this has been shown to occur spontaneously; however, if the cytosines are not turned out spontaneously, the HhaI Mtase enzyme can actively perform this mechanism if necessary (10). Upon binding to the DNA double helix, the enzyme HhaI MTase opens the C:G base pair. The Gln-237 residue in the active site of HhaI Mtase does this by sterically clashing with the cytosine, and stabilizing the unpaired guanine base. With the cytosine base pair now exposed, DNA methylation can occur more easily (11).
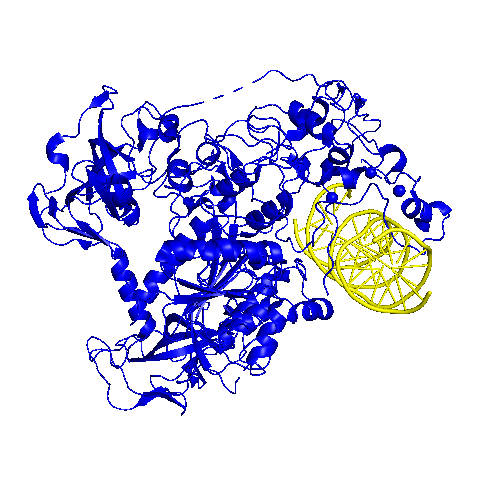
show the cytosine that is “flipped” out of the double helix by HhaI Mtase
(PDB-3PTA) (12).
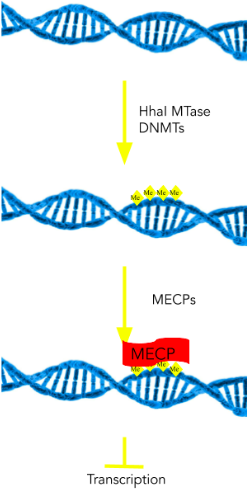
The key enzymes that performs the methylation of cytosines are DNA Methyltransferases (DNMTs), as shown above in Figure 6. When the cytosine is flipped out, the Glu-1266 residue helps stabilize the base in the active site, while Cys-1226 nucleophilically attacks the base (13). To see the mechanism, visit the DNA methylation mechanism page.
Once certain portions of DNA are methylated, gene expression can be altered. Methyl-CpG-binding proteins (MECPs) interact with methylated CpG islands, physically binding to the methylated areas of the DNA strand. Gene repression can occur in two ways: structural (MECP physically blocks transcription machinery) or enzymatic (recruits Histone Deacetylases, which help condense chromatin structure) (14). Interestingly, about 70% of promoter regions in DNA can be found in CpG regions, highlighting their role in gene repression (9).

This is clear and well done! The figures are really nice!
This page is very well defined and locations of methylations and where the DNA is actually methylated is all clearly demonstrated. It really helps add to the understanding on how exactly methylation results in the difference in transcription.
As a small correction: “All cells *of a eukaryote* contain the same genetic information…”
Other than that, everything is great!
Edited!
Is there anything that determines if the turning out of cytosines happens spontaneously or requires enzymatic help?
I think this whole page is very useful and I appreciate how even though the mechanism is linked to the other page, you still give a full view of what happens before and after this part to actually result in the gene repression.
I made some edits that will hopefully be helpful!