Epigenetic Inheritance of Diabetes
The increasing prevalence of cardiometabolic disease in the modern world has increased research efforts to elucidate the pathogenesis, especially that of type 2 diabetes. While researchers have extensively investigated environmental factors, especially calorie-rich diets and physical inactivity throughout the individual’s lifetime, there has been growing interest in epigenetic inheritance of diabetes.
GENERAL EXPERIMENTAL SETUP
Experiments in this area follow a general pattern. The epigenetic process of increasing the disease risk begins with the parent, the F0 generation. He or she faces environmental conditions that affect their glucose metabolism. To simulate these nutritional challenges, many experiments subject healthy F0 male or female mouse models to various conditions, such as high-fat feeding and low-protein feeding. [4] These conditions result in epigenetic modifications, such as DNA methylation and histone modifications, and subsequently altered gene expressions. [18] For example, overnutrition may upregulate genes that are involved in the metabolism of lipid biosynthesis, and undernutrition may downregulate genes affecting beta cell function. [4] Methylome analyses, such as MeDIP-Seq, often reveal that the epigenetic marks resulting from nutritional challenges may be transferred to the F0 germ cells.
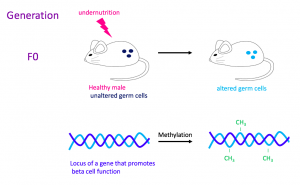
Figure 1. Starvation and its epigenetic effect. This illustrates an example of an experimental setup in the research on the epigenetic inheritance of risk of diabetes. A healthy male mouse is subjected to undernutrition, and the gene of interest encodes for the synthesis of an ion channel that allows insulin secretion. In germ cells, this treatment triggers the methylation of this gene, downregulating it. In this particular example, the treatment does not affect the metabolic phenotype of the F0 mouse, but other treatments, such as high-fat feeding, may lead to pre-diabetic symptoms.
The next step is to mate the F0 mouse, which now has an altered epigenetic profile, with a healthy one to breed the F1 generation. The metabolic phenotype and epigenetic alterations in F1 and F0 are compared. Due to the experimental setup, the risk of diabetes should be inherited only by epigenetic means, since neither of the F0 parents had the genetic predisposition to develop the disease, and only one F0 parent, not the F1 offspring, experienced the treatment. If the F1 offspring shows pre-diabetic symptoms, especially glucose intolerance and insulin sensitivity, then the epigenetic inheritance of the risk of diabetes may have taken place. Similarities between the epigenetic profiles of the F0 parent and F1 offspring further support this hypothesis. This page explains a recently performed experiment in which a mouse with induced pre-diabetes was mated with a healthy mouse, and then the methylome of its offspring was analyzed. [22]
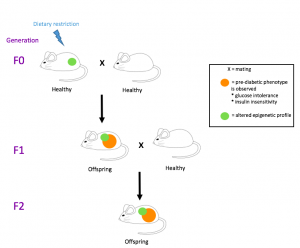 Figure 2. Common experimental design. (Adapted from ref. [4]) Research on the epigenetic inheritance of risk of diabetes follow a similar setup: subject a F0 mouse to a dietary restriction and mate it with a phenotypically normal mouse. The F0 may be male or female. Depending on the treatment, the F0 mouse may or may not develop a pre-diabetic phenotype. The metabolic phenotypes, especially glucose tolerance and insulin sensitivity, as well as the epigenetic profile of the F1 and F2 offspring are analyzed.
Figure 2. Common experimental design. (Adapted from ref. [4]) Research on the epigenetic inheritance of risk of diabetes follow a similar setup: subject a F0 mouse to a dietary restriction and mate it with a phenotypically normal mouse. The F0 may be male or female. Depending on the treatment, the F0 mouse may or may not develop a pre-diabetic phenotype. The metabolic phenotypes, especially glucose tolerance and insulin sensitivity, as well as the epigenetic profile of the F1 and F2 offspring are analyzed.
BARRIERS TO EPIGENETIC INHERITANCE
The epigenetic inheritance of any phenotype depends on the stability of the alterations. After all, epigenetic reprogramming takes place during gametogenesis and embryogenesis — TET enzymes demethylate regions of the genome, and protamines replace parental histones. [4] The removal of these epigenetic marks during reprogramming makes their presence in the F1 and F2 offspring highly unlikely. Therefore, biochemical pathway must be preventing the demethylation and histone replacement at certain loci of the genome and preserving the epigenetic marks on the offspring.
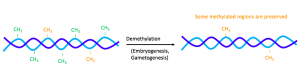 Figure 3. Demethylation during gametogenesis and embryogenesis
Figure 3. Demethylation during gametogenesis and embryogenesis
Research on the molecular mechanism that stabilizes the epigenetic marks in mammals is relatively new. [19] The major breakthrough has been the discovery of factors that bind to histones, preventing the TET enzymes from demethylating imprinted loci, and this page summarizes the experiment. [19]
November 12, 2017 at 8:25 pm
I like the description of experimental methods used to explore epigenetics.
For the barrier section, I think it’d be cool to see if any modifications are more stable than others, thus having a higher chance of inheritance. In addition, I assume that it’s a collection of various modifications that will result in higher chance of development of diabetes; are there any modifications that will influence disease development more than others?
Research on this may also be nonexistent so that may be impossible.
November 12, 2017 at 8:28 pm
I forgot to include this in the original comment, but sometimes terms are introduced that are somewhat confusing. For instance, the first time “insult” was used in the description of figure 1, I thought that insult was a misspelled insulin. I think it’d be helpful to define these words before using them.
November 13, 2017 at 8:49 pm
Building off of what Frank commented, have any trends arisen from the experiments described from this section, or is this field of study still in the conceptual stage? Similarly, how much research has been conducted on the factors blocking TET activity?
Also, this may not be necessary to include in the website, but a short explanation on how epigenetic profiles are taken and compared may be good for a better overall picture of how experiments and research are conducted.
November 13, 2017 at 8:54 pm
Just read through some of the recent research pages, linking or referencing some of the more common methods used might clear up some of the stuff I mentioned before.