Epigenetics in Molecular Detail
EPIGENETICS: Histone Modification
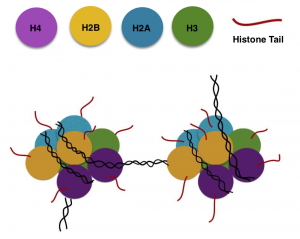
Figure 1. Histone, the octameric complex with H2A, H2b, H3 and H4
Adapted from Kim, 2014 [6]
DNA in eukaryotic cells are arranged with proteins called histones—this octameric complex is called nucleosome and composed of two molecules of each of the 4 histone types: H2A, H2B, H3 and H4, as Figure 1 shows. [1], [2] The NH2 terminal tails of the histones are modifiable by acetylation, methylation, phosphorylation, sumolyation, ubquitination, and many more. [1], [3]
The most common histone modifications are acetylation and methylation of the lysine residues in the N termini of H3 and H4. [2]
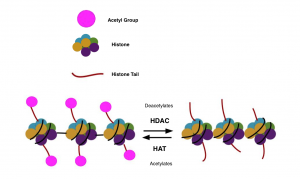
Figure 2. HDAC and HAT, removal and addition of acetyl groups to histone tails
Adapted from Edwards & Pender, 2011 [7]
Histones Deacetylases (HDACs) remove and histone acetyl transferases (HATs) add acetyl groups to the lysine part of the histone tails, as seen in Figure 2. [1] Methylation of histone affects gene activity in that K4 methylation of H3 will increase gene activity while methylation of K9 of H3 will decrease gene activity. [3]
EPIGENETICS: DNA methylation
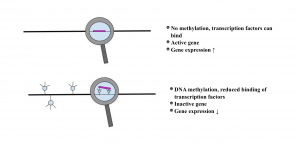
Figure 3. DNA methylation and its effect on on transcription
Adapted from Ling & Groop 2009 [1]
DNA methylation is often correlated with transcriptional silencing. The silencing can be brought upon by prohibiting the transcription factor binding or by engaging proteins that bind to the methylated CGs that consequently engage histone deacetyltransferases (HDACs) and corepressors. [1]
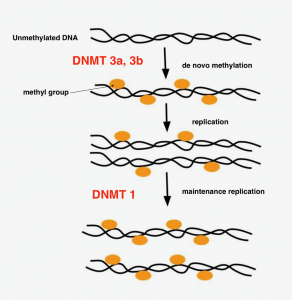
Figure 4. DNA methylation by DNA methyltransferases (DNMT)
Adapted from ATDBio [10]
DNA methylation is driven by DNA methyltransferases (DNMT). There are three major types of DNMTs: DNMT 1, DNMT 3a and DNMT 3b. [3] DNMT1 is often associated with DNA methylation between cell generations during DNA replication (maintenance) while DNMT 3a and 3b is associated with de novo methylation, which means new 5-methylcytosines are introduced to DNA that were not previously methylated. [1] , [3] In addition to the three major types of DNMTs, there is also a regulatory factor DNMT3L that takes part in DNA methylation. In general terms, gene activity will be down regulated when DNA is methylated, as Figure 4 demonstrates. [3]
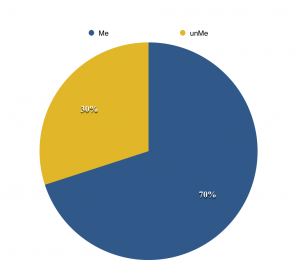
Figure 5. CpG dinucleotide methylation
Data from Pinney & Simmons, 2009 [2]
Figure 6. CpG island, CpG dinucleotide
Cytosine base is modified by DNMT at C5, mostly within CpG dinucleotides. [2] , [3] About 70% of CpG dinucleotides in human DNA are methylated and the unmethylated CpGs are mainly located at CpG islands (Figure 5). CpG islands are CG-rich sequences that operates as a promotor to determine the affinity of transcription factors to the binding sites on DNA. While many normal cells have their CpG islands unmethylated, with cancer and oxidative stress, they become methylated de novo and is often brought upon with histone modifications to make repressed gene transcription (Figure 6). [2]
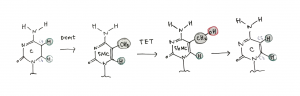
Figure 7. DNA cytosine methylation, hydroxylation, and demethylation
Redrawn from Tollefsbol, 2011 [3]
DNMT catalyzes the transfer of methyl part from S-adenosyl-L-methinione (SAM) to the C5 of cytosine in particular CpG dinucleotides (Figure 7). Mtase binding to the DNA, the targeted nucleotide does a outward turn so that the targeted part projects out of the double helix (“base flipping”) and there is a covalent attack of the Cys nucelophile on the cytosine C6, transfer of methyl group from SAM to the activated cytosine C6. [3] Detailed mechanism can be found under Pathways and Mechanism page, at the “DNA Methylation Mechanism” tab, or Here.
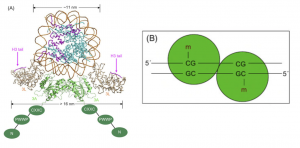
Figure 8. DNA binding domain dimerization.
8A. Nucleosome with Dnmt3L-3a-3a-3L tetramer 8B. Dnmt3a dimer
Directly adopted from Tollefsbol, 2011 [3]
The active site of DNMT 3a and 3b have one of the smallest DNA binding domains in midst of all DNA MTases. By dimerization, such as 3a-3a and 3b-3b, the active sites are brought together to double the binding area. The two actives sites are located in the major groove of DNA and the dimeric DMNT3a allows for methylating two CpGs separated by one helical turn in one binding event. [3] (Figure 8)
To get a summarized overview, watch this video! Below are the time stamps of the video for guidance.
0:00-0:21 Introduction
0:21-1:30 Histone, Nucleosome, DNA Sequence
1:30-2:09 DNA & Histone Methylation
2:09-2:26 Epigenetics Overview
2:26-2:51 Introduction to Nucleosome, Histone Tails
2:51-3:25 DNA Methylation, CpG sequence
3:25-3:50 Histone Modification, Acetyl and Methyl groups addition
3:50-4:09 Conclusion
Video 1. Epigenetics – An Introduction [5]

November 12, 2017 at 6:55 pm
I like that the text is separated by figures which help to clarify the information presented. There are a couple typos here and there, but overall a pretty solid page.
I think you could kind of combine this page with the next page that explores mechanisms of each modification by linking the pages on this page as well.
November 12, 2017 at 8:09 pm
I’m not sure if this possible, but I think it would also be helpful to explain which types of protein expression are upregulated/downregulated in response to the methylation.
It would also be cool to explore more about what acetylation does to gene expression if possible because half of the pathways is on acetylation/deacetylation.
November 13, 2017 at 5:31 pm
Using the video to give a quick general overview was a cool idea. The visuals on this page, as Frank said, are very helpful in explaining their respective topics. I had a bit of trouble understanding the section starting with “Cytosine base is modified by DNMT at C5…”. At the beginning of the paragraph it is stated that 70% of CpG islands are methylated. Towards the end of the same paragraph, however, the website states that “many normal cells have their CpG islands unmethylated”.