What is a structured search?
A structured search is a systematic approach to finding references in a literature database using Boolean operators (AND, OR, NOT), keywords and controlled vocabulary terms, such Medical Subject Headings (MeSH). The goal of a structured search strategy is to balance recall and relevance (sensitivity and specificity).
How do I create a structured search?
The key to creating a good structured search is doing a little work before you go to a database.
Step 1: Develop a focused question
Like all research, a good search begins with a good question. Health professional students are taught to use the acronym PICO to construct clinical questions, where ‘P’ stands for patient or problem, ‘I’ for intervention, exposure or prognostic factor, ‘C’ for comparison, and ‘O’ for outcome. The idea is to create a well-defined question with multiple concepts, which helps you build a search strategy and evaluate the relevance of your search results. You may need to modify your question once you conduct a few searches.
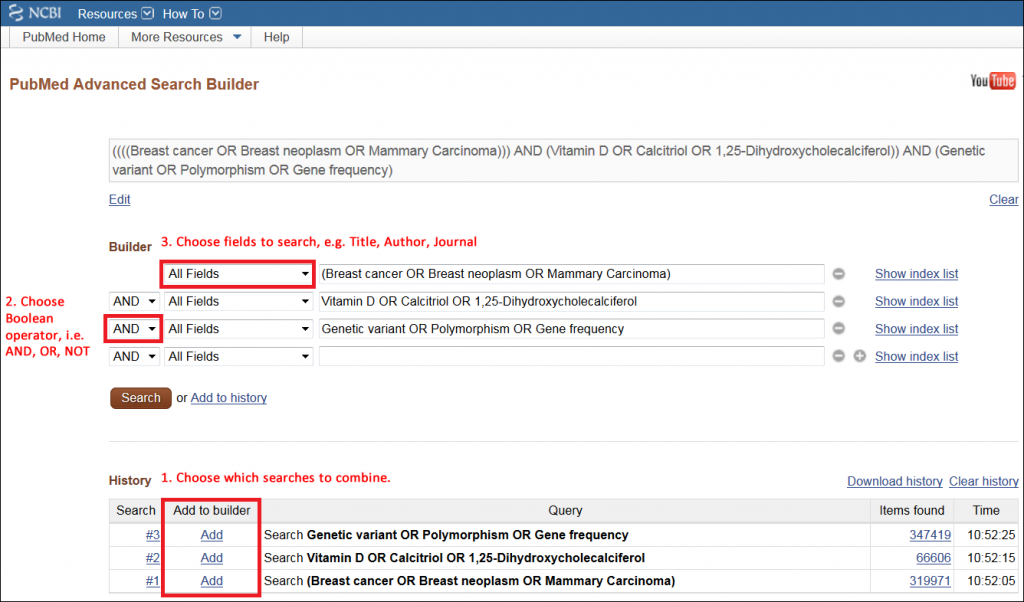
Example: How do genetic variants in the vitamin D pathway affect breast cancer risk?
Step 2: Identify the key concepts of your questions.
Break down your question into its components. You can use the PICO acronym, or simply think: who, what, when, where, how.
Example: How do genetic variants in the vitamin D pathway affect breast cancer risk?
Breast cancer (concept 1)
Vitamin D (concept 2)
Genetic variants (concept 3)
Step 3: Choose keywords and standardized (controlled vocabulary) terms to describe each concept.
The goal of this step is to think of different ways to describe each concept. Keywords are natural language, the terms you use when discussing the concept with a colleague; consider acronyms, abbreviations and close synonyms. Standardized terms are from a controlled vocabulary, such as MeSH in PubMed; not all databases have a controlled vocabulary. The inclusion of multiple keywords and standardized terms ensures that you do not miss relevant articles on your topic, regardless of how an author or indexer described the topic.
Example: How do genetic variants in the vitamin D pathway affect breast cancer risk?
Breast cancer (concept 1): Breast neoplasms, mammary carcinoma…
Vitamin D (concept 2): Calcitriol, 1,25-dihydroxycholecalciferol…
Step 4: Using Boolean operators, search each concept separately then combine.
Boolean operators (AND, OR, NOT) are used to combine words and phrases in a search strategy. Use ‘OR’ to combine all keywords and standardized terms for one concept and run this search in a database. Once you have searched each concept separately, then combine different concepts using ‘AND’. Searching each concept separately allows you to identify any problems with particular terms before you build a complicated search, and gives you the flexibility of combining your concepts in different ways.
Example: (Breast cancer OR Breast neoplasm OR Mammary Carcinoma) AND (Vitamin D OR Calcitriol OR 1,25-Dihydroxycholecalciferol) AND (Genetic variant OR Polymorphism OR Gene frequency)
| OPERATOR |
SEARCH |
RESULTS |
VENN DIAGRAM |
| AND |
Heart disease AND hypertension |
Articles containing BOTH heart disease and hypertension |
 |
| OR |
Heart disease OR Hypertension |
Articles containing EITHER heart disease, hypertension, or both |
 |
| NOT |
Heart disease NOT hypertension |
Articles containing ONLY heart disease, not hypertension |
 |
Step 5: Use filters to limit results.
Most databases have filters, such as date, language and publication type, that allow you to further narrow your results. Be judicious when using filters. If you have too many irrelevant results, then you need to modify your search, not apply more filters.
This sounds complicated and time-consuming, do I really need to construct a structured search each time I need to find articles?
Not necessarily. If you just need a few good articles, then you can enter a couple terms in a database and scan the results. However, if you are doing a literature search for your dissertation, qualifiers, or grant proposal, then it is a good idea to do a structured search. This strategy may require an initial investment of time, but it will (hopefully) save you the frustration of scrolling through hundreds of irrelevant results, or missing an important article. Of course, I am always available to help you construct a search strategy.