Bioinformatics
Abstract
Bioinformatics is the use of information with computation for problem solving in various fields of biology. This field has experienced tremendous growth since the early 2000’s at the start of The Human Genome Project and has been used heavily in proteomics and genomics. The use of FPGA’s is a promising way to increase computing speed, and computer tools such as BLAST and iBIRA make the ever-growing amount of biological data more manageable, but computer scientists and engineers must continue to find solutions. Due to the continually growing nature of the field, the need for increased bioinformatics education in school is addressed.
Introduction
Bioinformatics is a broad field that lacks a concrete definition as it encompasses elements of many other fields. It is a way to study biology by applying techniques from computer science and statistics on large amounts of data (Luscombe, Greenbaum, & Gerstein, 2001). As more data becomes available in the biological field and the sciences in general, the need for computers to process and analyze all of this data has also grown. Computers can do in seconds what it would take humans to do in hours, making the use of computers in the field of biology a near necessity.
Bioinformatics is a branch of informatics, with other branches including fields such as construction, geology, and medicine. In its simplest form, informatics is considered as the “science of data with meaning” (Bernstam, Smith, & Johnson, 2010). Bioinformatics is adding meaning to data, but with a focus in biology.
This article aims to give the reader a high-level understanding of bioinformatics. The main uses of bioinformatics are discussed followed by the importance of engineering. Challenges and solutions to computer limitations, computer science, and two software packages that are being used today are covered, as well as the similarities between bioinformatics and computational biology. Finally, there is a discussion on including bioinformatics education in school.
The Main Uses of Bioinformatics
While bioinformatics was being used before the turn of the century, it really came into the public eye in 2003 upon completion of the Human Genome Project. The aim of this research initiative was to find and store the sequence of the three billion nucleotides that make up human DNA for research in creating medicines tailored to the individual. With this much data, there needed to be a sophisticated way of storing and analyzing it in a reasonable amount of time (Ram, Sureka, Sharma, & Rao, 2010). Even prior to this research, the GenBank repository of nucleic acid sequences had 11,546,000 entries and the SWISS-PROT database of protein sequences had 95,320 entries – values much too large to be analyzed by hand. These numbers are from the early 2000’s and continue to double in size every fifteen months (Luscombe, Greenbaum, & Gerstein, 2001). Engineering is not only greatly needed to satisfy the computing needs of today, but must also keep up in the fields of biology that have come to depend on computers.
The two main uses of bioinformatics over the last fifteen years have been in proteomics, the study of proteins, and genomics, the sequencing, assembling, and analyzing of the complete set of DNA of an organism (Rojas, Pomares, Valenzuela, & Bernier, 2009). It’s been used in bioinorganic chemistry to help identify and classify metalloproteomes, which are proteins that contain a metal, and in genetics and molecular biology to study patterns and the sequence of DNA (Bertini & Cavallaro, 2009; Chen & Skogerbø, 2010). A common misconception is that bioinformatics is directly related to the study of medicine and health. While there are applications of bioinformatics that have been focused in the medical field, in general it pertains to the advancement of biological research through computing, like the Human Genome Project.
The inclusion of bioinformatics in the fields of proteomics and genomics has helped speed up growth and research. But there are still challenges on the engineering side that must be solved if the field is to continue being beneficial.
Computing Challenges and a Potential Solution
While bioinformatics has been very helpful for biological research, there are limitations on the computer and engineering side. The algorithms needed demand a lot of computing power not due to complexity of code, but because of the massive amount of information that is being processed (Pfeiffer, Baumgart, Schröder, & Schimmler, 2009). Only fourteen years ago, computers were already processing a tremendous amount of biological data as it pertained to human DNA: 12.5 billion nucleotide bases in DNA sequencing, 120 million amino acids in protein sequences, and another 300 genomes with 1.6 – 3 billion nucleotide bases each (Luscombe, Greenbaum, & Gerstein, 2001). It is simply unreasonable to process billions of pieces of data without computers. The size of DNA sequence databases doubles twice a year, whereas computing performance doubles only once every two years (Pfeiffer, Baumgart, Schröder, & Schimmler, 2009). There is a serious need for improved computing power in order to keep up with demand.
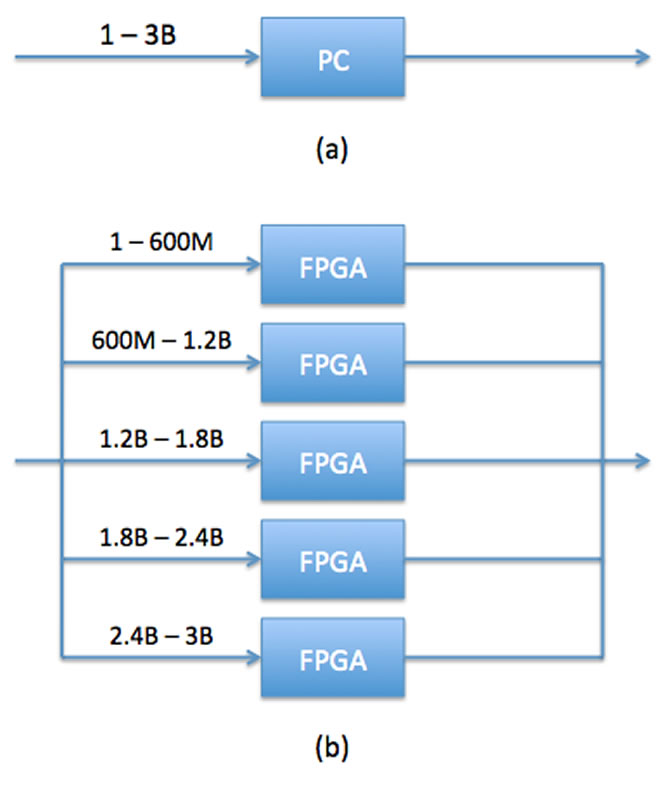
One proposed method of increasing computing speed while keeping cost low is to use many FPGA’s simultaneously (in parallel) as opposed to one after another (in series). An FPGA is a Field Programmable Gate Array. These chips can be programmed after they are manufactured to accomplish whatever task the user has in mind (within the limitations of the chip). By using a number of FPGA’s in parallel, the same task that a single computer processes can be done by a number of machines all at the same time, thus increasing speed (Figure 1).
Figure 1
(a) depicts how a sequence of 3 billion nucleotide bases in a strand of DNA would be processed in a single PC. In (b) there are five FPGA’s, so each one can process a specified number simultaneously, therefore saving time. Source: Wiegard (2015).
A Focus on Computer Science
Simply by definition, computer programming and engineering is a large part of bioinformatics. However, most of the engineering involved is related to computers. While it is not specific to bioinformatics, electrical and computer engineering is important in developing servers and processors for increases in computing speed and power. These aspects need to be addressed as more data becomes available, but the features that biologists use are largely a product of computer science. It should be noted that computer science and computer engineering are distinct, which makes it important for those dealing with the computing side of bioinformatics to understand both. Computer science, in this case, represents the creation of the organizational and analytical algorithms and tools that are used to better understand the information being studied. Computer engineering is concerned with things such as memory, computing speed, and the overall cost of the system in a physical sense.
Computer scientists are equally as important in bioinformatics as the biologists doing the research. They are the ones creating the algorithms that allow biologists to analyze their work at a fast pace. Here we mention two different kinds of software that demonstrate the power of bioinformatics.
BLAST
One of the more widely recognized software tools in bioinformatics is called Basic Local Alignment Search Tool (BLAST) (Cha & Rouchka, 2005). This tool can select a newly found gene in an animal and compare it to the human genome to see if humans carry a similar gene. BLAST will automatically compare the sequence from the animal and find ones that are similar in the human sequence. This software is very useful as these sequences can be hundreds or even thousands of bases long.
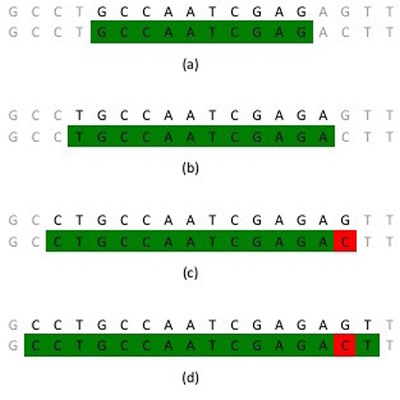
Computer science manages this data. In a lot of cases, very long sequences do not match entirely. Therefore, it does not make much sense to see if, for example, one thousand sequential bases match exactly with another one thousand sequential bases. Instead, the computer scientists who developed BLAST* designed it such that it first only compares short sequences (around 10-20 bases) to see if anything matches with 100% accuracy. BLAST then takes these matches and extends outwards to determine how many bases match. It can even skip over a few bases to see if the similar sequence continues after being different for a short period (Cha & Rouchka, 2005). Figure 2 demonstrates this process.
Figure 2
Example strand of DNA illustrating how BLAST works. The two rows of DNA are from separate organisms. BLAST starts with a short sequence (a) and extends outwards one nucleotide base (b). It extends again (c), takes note of a difference in the two sequences, and continues on (d). Source: Wiegard (2015).
The BLAST algorithm shows a glimpse of how important computer science is in bioinformatics. Without it, biologists would be forced to manually compare these long genome sequences, which is not only inefficient, but opens the door for mistakes. Computer scientists designed this automated process by breaking down the task piece by piece, just as any good computer scientist would do. They took the time to consider how the program was going to be used, which is very important, because if the person who is actually using the program cannot do so efficiently or effectively, then the program is unhelpful.
iBIRA
Another tool designed to increase simplicity is the Integrated Bioinformatics Information Resources Access (iBIRA). It is aimed to be the all-seeing power in bioinformatics, with duties including developing and coordinating access to other databases and resources, finding cross references, and providing information to users (Ram, Sureka, Sharma, & Rao, 2010). Information from many different sources gets put onto this large database and iBIRA takes care of organizing the information in groups that are determined by the user.
iBIRA is like an intelligent Wikipedia for bioinformatics. Those who wish to share their knowledge and findings can put information online, and iBIRA automatically categorizes it (for example, into sections for journals, institutions, and bio-companies), allows for easy searching, and can locate similar topics to what the user is viewing. A lot of computer science and engineering had to be used in this task in order to not only develop the categorization and searching methods, but also in the storage and protection of this database.
A Note on Security
Topics such as storage, power, and speed are important in bioinformatics, but equally as important is the security of information. While most of the time the information of focus is not fully online, it could exist on a network of computers in a single research lab. When valuable information is stored on a computer, there is always opportunity for it to be stolen. Computer scientists and engineers must take security into account when creating a storage database. The challenge is do so in a way that makes it relatively easy for the true users of the system to access their information without having to get through layers and layers of security every time they log in.
Bioinformatics is not the only field with reliance on computing, however, and some of the aspects discussed here were needed before the Human Genome Project was completed. Computational biology is another field that has been positively affected by the use of computers.
Computational Biology
A more common connection between computers and biology is computational biology. This field is defined as “the development of techniques for the collection and manipulation of biological data, and the use of such data to make biological discoveries or predictions” (MeSH Browser). While there is still debate on whether this field is differing in scale to, similar to, or the same as bioinformatics, the definition is quite similar to the broad definition of bioinformatics – applying techniques on large amounts of biological data to better understand it.
Computational biology is flooded with a large amount of complex data and thus has a need for organized, accessible, and secure storage. In the late eighties and early nineties, algorithms existed for analyzing blurry images taken with an electron microscope. But until there were increases in computational speed in the mid-nineties, the practical implementation of such algorithms was impossible (Kingsbury, 1996).
While there has been great progress in computational biology and bioinformatics over the last two decades, one area that is still not getting enough attention is education and exposure in an undergraduate program.
Bioinformatics in Schools
Bioinformatics is a very interdisciplinary field that requires more than one traditional education curriculum to become truly knowledgeable. Very little exists in terms of formal education in bioinformatics. Most universities teach biology and/or computer science and engineering and offer one or two courses in bioinformatics, but few offer full coverage of the combination of both science and computing.
Those who work in bioinformatics today typically come from one of these two educational backgrounds, and biologists and computer scientists work together to create the databases and organization that makes this field so useful. However, it would be beneficial to have individuals that not only understand the computing required, but also are experienced in the biological sciences, or vice versa. In a study at Tuskegee University, thirty-nine life sciences students were given a questionnaire having to do with computers. On a scale of 1-5, with 5 representing “most favorable”, the average response to the question “Do you agree that better computer skills can help your career?” was a 4.9. But when asked if they knew what Linux is (a computer operating system, like Windows) and what a p-value is (a measurement of probability in statistics), only 15% and 23% answered correctly, respectively (Qin, 2009). So the understanding of its importance is there, but the actual understanding is not.
Having an individual who understands both the computer and biology aspects helps to minimize the opportunities for misunderstandings. For example, perhaps a programmer knows of a great way to store and analyze the sequence of human DNA by maximizing speed and minimizing memory space needed. However, suppose this method makes it very difficult for a biologist to do their work because it is hard to access or it is too complex to use. By having an individual who understands the needs and limitations of both sides, perhaps more widely accepted and useful solutions can be achieved (Umarji, Seaman, Koru, & Liu, 2009). This type of education would have its benefits, and it all starts by getting exposure as an undergraduate.
Proposed Solutions
One proposed method for introducing bioinformatics into an undergraduate education is not to create an entirely separate program, but to introduce it in small segments into courses that already exist. A database course could incorporate a lesson on biological databases and a biology course could teach computing methods when analyzing DNA patterns. (Poe, Venkatraman, Hansen, & Singh, 2009). Entirely new courses would result in a lot of overlap of material that courses in statistics, computer engineering, and biology already cover. By introducing bioinformatics topics early on, it helps lay the foundation and spark interest for undergraduates to pursue this field in their future studies.
Conclusion
The field of biology is growing each day and technology must keep up if it is to stay that way. Bioinformatics has been very useful for research in subsets of biology such as proteomics and genomics and its future looks bright. There is a clear need for computer scientists and engineers in this field to keep up with demand and it is not going away any time soon. Implementation of bioinformatics software has been successful thus far and educating today’s youth in this relatively new field will only help to continue solving problems and furthering biological research.
The developers are Stephen Altschul, Warren Gish, Webb Miller, Eugene Myers, and David J. Lipman at the NIH.
Bibliography
- Bernstam, E. V., Smith, J. W., Johnson, T. R. (2010). What is biomedical Informatics. Journal of Biomedical Informatics, 43(1), 104-110. DOI: http://dx.doi.org/10.1016/j.jbi.2009.08.006
- Bertini, I., Cavallaro, G. (2009). Bioinformatics in bioinorganic chemistry. Metallomics, 2(1), 39-51. DOI: http://dx.doi.org/10.1039/b912156k
- Cha, I.E., Rouchka, E.C. (2005). Comparison of current BLAST software on nucleotide sequences. 19th IEEE International Parallel and Distributed Processing Symposium. DOI: http://dx.doi.org/10.1109/IPDPS.2005.145
- Chen, R., Skogerbø, G. (2010). Bioinformatics – Mining the Genome for Information. Frontiers of Electrical and Electronic Engineering in China, 5(3), 391-404. DOI: http://dx.doi.org/10.1007/s11460-010-0109-8
- Kingsbury, D.T. (1996). Computational Biology. ACM Computing Surveys, 28(1), 101-103. DOI: http://dx.doi.org/10.1145/234313.234358
- Luscombe, N. M., Greenbaum, D., Gerstein, M. (2001). What is bioinformatics? A proposed definition and overview of the field. Methods of Information in Medicine, 40(4), 346-358. Retrieved from http://www.binf.gmu.edu/vaisman/binf630/mim01-luscombe-what-is-bioinf.pdf
- MeSH Browser [Internet]. Bethesda (MD): National Center for Biotechnology Information (US), Computational Biology; [cited 2015 Jan 28]; Retrieved from http://www.ncbi.nlm.nih.gov/mesh/?term=bioinformatics
- Pfeiffer, G., Baumgart, S., Schröder, J., Schimmler, M. (2009). A Massively Parallel Architecture for Bioinformatics. Lecture Notes in Computer Science, 5544,994-1003. DOI: http://dx.doi.org/10.1007/978-3-642-01970-8_100
- Poe, D., Venkatraman, N. Hansen, C., Singh, G. (2009). Component-Based Approach for Education Students in Bioinformatics. IEEE Transactions on Education, 52(1), 1-9. DOI: http://dx.doi.org/10.1109/TE.2007.914943
- Qin, H. (2009). Teaching computational thinking through bioinformatics to biology students. SIGCSE, 41(1), 188-191. DOI: http://dx.doi.org/10.1145/1508865.1508932
- Ram, S., Sureka, R. K., Sharma, P., Rao, N. L. (2010). Integrating Bioinformatics Information Resources: Management of Information Through Clustering. IEEE Students’ Technology Symposium. 177-180. DOI: http://dx.doi.org/10.1109/TECHSYM.2010.5469176
- Rojas, I., Pomares, H., Valenzuela, O., Bernier, J.L. (2009). Applications in Bio-Informatics and Biomedical Engineering. Bio-Inspired Systems: Computational and Ambient Intelligence, 5517, 820-828. DOI: http://dx.doi.org/10.1007/978-3-642-02478-8_103
- Umarji, M., Seaman, C., Koru, A. G., Liu, H. (2009). Software Engineering Education for Bioinformatics. 22nd Conference on Software Engineering Education and Training. 216-223. DOI: http://dx.doi.org/10.1109/CSEET.2009.44
Suggested Reading
- GenBank Repository. Retrieved from http://www.ncbi.nlm.nih.gov/genbank/
- iBIRA (Integrated Bioinformatics Information Resource Access). Retrieved from http://ibiranet.in/
- Informatics. (n.d.). In NCBI. Retrieved from http://www.ncbi.nlm.nih.gov/mesh/68048088
- IEEE/ACM Transactions on Computational Biology and Bioinformatics. Retrieved from http://ieeexplore.ieee.org/xpl/RecentIssue.jsp?punumber=8857