Outreach: Peptide MD Simulation
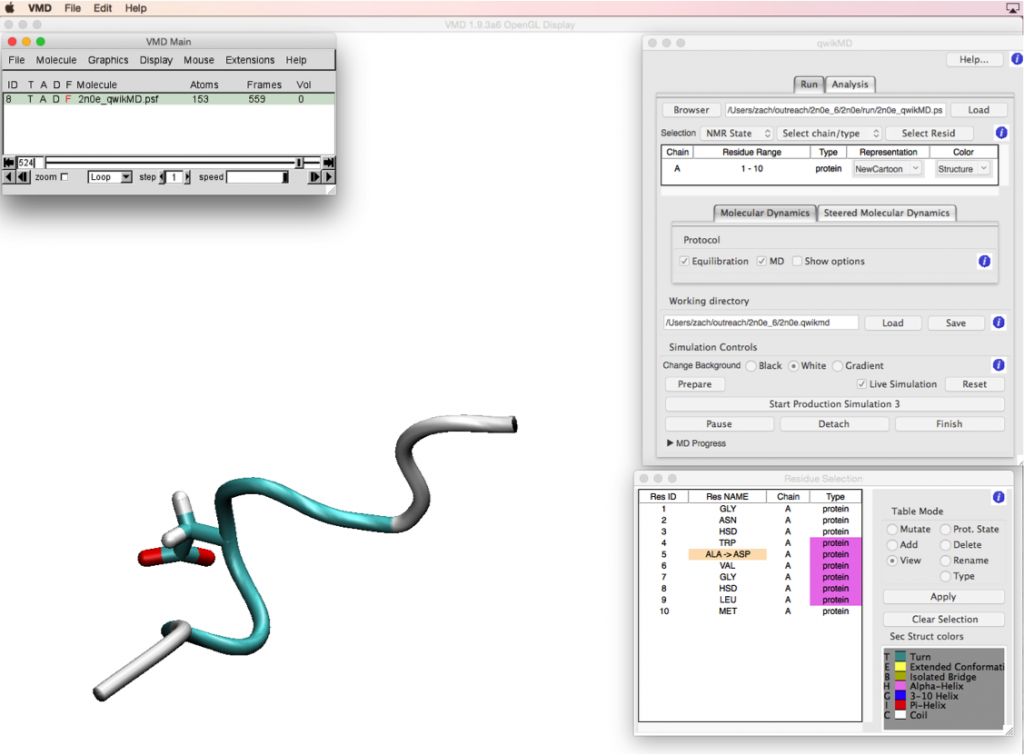
In this module we demonstrate how the amino acid sequence of a protein dictates its structure, and how molecular dynamics (MD) simulations can be used to study protein dynamics and their sequence‒structure relationships. In this module, the QwikMD plugin in VMD is used to run simulations of a small helical peptide. As the simulation runs, students see a GUI window updating on the fly to show the structural changes the peptide undergoes. Students then apply what they have learned about amino acids and factors that stabilize protein structures to estimate the extent to which a mutation might influence the structural stability of the peptide. Students also learn how MD simulations work and how they can be used to study protein dynamics, as well as how to actually set up MD simulations using QwikMD and mutate amino acids at a given position. They can then run their own QwikMD simulations to investigate how mutations affect the native structure of the peptide and discuss the simulation results compared to their original hypothesis.
MA State Standards Addressed:
9-12.DTC.a.1, 9-12.DTC.b.1, 9-12.CT.e.1, 9-12.CT.e.2, HS-LS1-1, HS-LS1-6, HS-LS4-4
We’d love to know about any questions, issues when implementing this module, and feedback you have! Please email your comments and questions to YSL.
