Outreach: Docking Simulation
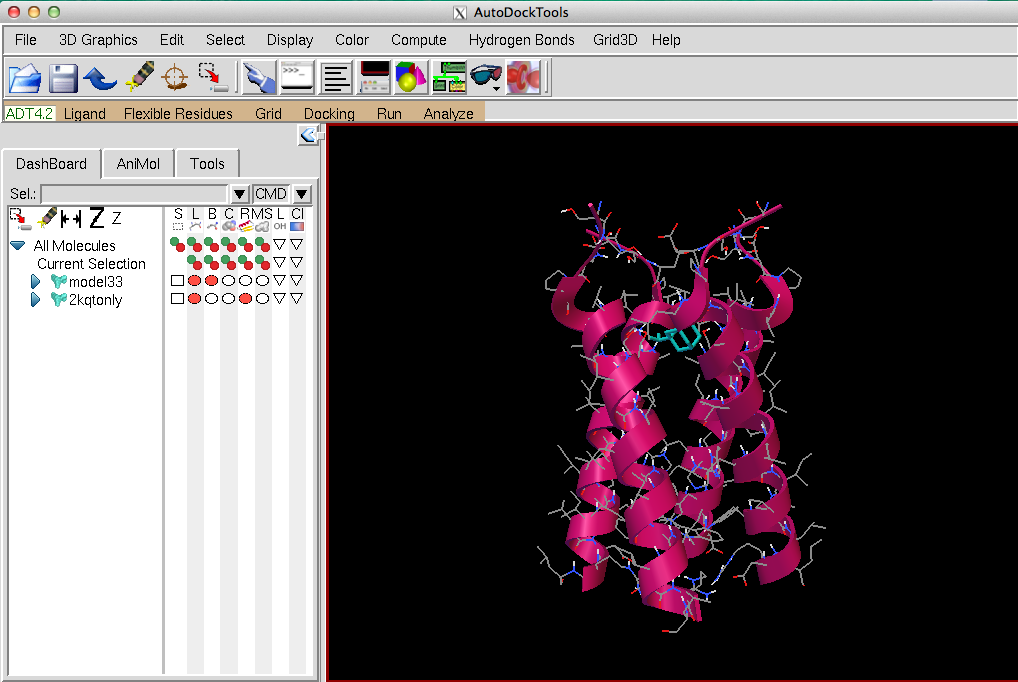
In this module students will learn how viruses develop drug resistance, and how docking simulations can be used to estimate binding affinity of a compound to a protein. Students learn how docking simulations work and perform docking of a discontinued antiviral drug, amantadine (shown in cyan in the figure below), on the wild-type M2 proton channel (shown in pink ribbon). Students examine the binding of amantadine to the wild-type M2 proton channel and hypothesize what kinds of mutations on which amino acids might lead to drug resistance. Finally, students perform a docking simulation of amantadine to the known drug-resistant S31N M2 proton channel. In this docking simulation students fail to obtain any binding pose with large binding affinity and thus confirm the resistance of this mutant M2 channel to the discontinued amantadine.
MA State Standards Addressed:
9-12.DTC.a.1, 9-12.DTC.b.1, 9-12.CT.e.1, 9-12.CT.e.2, HS-LS1-1, HS-LS1-6, HS-LS4-4
We’d love to know about any questions, issues when implementing this module, and feedback you have! Please email your comments and questions to YSL.
