Research
TB continues to kill millions of people every year. Mycobacterium tuberculosis remains the second deadliest infectious agent in the world. Our research focuses on understanding how mycobacteria tolerate stress and perturb host cell biology to evade killing by antibiotic treatment and the host immune response. We integrate single-cell measurements and computational modeling to quantitatively describe stress tolerance and virulence mechanisms of mycobacteria. We aim to use this knowledge to engineer improved therapeutics for treating tuberculosis. The complexity of disease and treatment challenges is not unique to TB, so approaches to optimize multidrug therapies directed against tuberculosis are expected to contribute greatly to rationally designed therapies for other heterogeneous and naturally drug-resistant infections and cancers.
Phenotypic mycobacterial drug tolerance:
Using live-cell microscopy and quantitative image analysis, we found that mycobacteria exhibit an asymmetric growth pattern that deterministically generates closely related cells with different growth properties and tolerance to drug treatment. We continue to use live-cell microscopy and computational modeling to quantify the relative contributions of key mycobacterial cell physiological properties on the ability of distinct subpopulations to tolerate different stressors. We also study how cell cycle progression and growth heterogeneity are mediated. Based on observations that cell morphologies change dramatically during drug treatment, we have developed an experimental and analysis pipeline to determine drug mode of action based on morphological features.
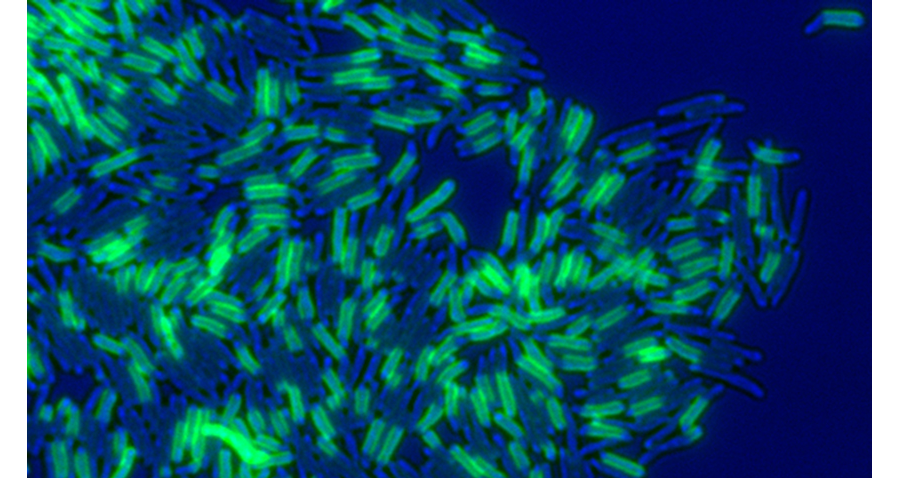 Figure 1. Mycobacterium smegmatis cells pulse-labeled with a fluorescent (green) amine-reactive dye, marking old cell wall material. The bright-field image is pseudo-colored blue.
Figure 1. Mycobacterium smegmatis cells pulse-labeled with a fluorescent (green) amine-reactive dye, marking old cell wall material. The bright-field image is pseudo-colored blue.
Design of combination therapy:
TB must be treated with combination therapy, and importantly, many organizations are now working together to facilitate collaboration in tuberculosis drug discovery. Our lab uses DiaMOND, a geometric optimization of traditional drug combination assays, to efficiently measure drug interactions and combination efficacies. We aim to use systematic measurement of drug combinations to optimize TB drug regimens and learn how to construct improved combinations. Our methods are broadly applicable to other disease models.

