EXPLOITATION OF CELLULAR PATHWAYS BY HERPESVIRUSES AND INFLUENZA A VIRUS TO RE-PROGRAM HOST GENE EXPRESSION AND INNATE RESPONSES
Viruses employ multiple factors to take over the infected cells, with the goal of repurposing resources and avoiding immune system detection. We want to unravel the role of host gene regulation in this viral take-over – we are working to uncover how viruses remodel the gene expression program of the host, which factors they use to do so and how the viral replication cycle and/or pathogenesis is affected by host gene regulation. Current projects in our lab include:
Viral RNases in host gene regulation (in collaboration with Craig McCormick, Denys Khaperskyy and Seema Lakdawala)

Regulation of RNA stability is often used by cells to fine-tune gene expression, especially of highly regulated genes. In contrast, several viruses hijack RNA degradation pathways to enact “host shutoff”, a wide-spread reduction in host gene expression, using virus-encoded ribonucleases (RNases). Viruses that encode host-shutoff RNases include gamma-herpesviruses like KSHV, alpha-herpesviruses, coronaviruses and orthomyxoviruses like influenza A virus. The RNases these virus encode are not molecularly related, but surprisingly share a similar mechanism to degrade host RNAs that involves host-encoded RNA degradation enzymes, essentially tricking the cell into degrading its own transcripts. We also found that, the action of these RNases is targeted in two different ways: 1) they specifically target transcripts generated by the RNA polymerase II complex; 2) in the case of the KSHV protein SOX, cleavage occurs at specific sites defined by a degenerate sequence/structural element; in the case of influenza A virus PA-X other determinants may determine the position of cleavage sites. This suggests that there is more to host gene regulation by viral RNases than currently understood. Questions we are working on include: 1) how is the specificity of these enzymes is achieved?; 2) how is this specificity used by the viruses to sculpt gene expression and affect host pathways? In particular, we want to unravel the mechanism of action of the influenza A virus RNase PA-X, which has a key role in the regulation of immune-related damage during influenza A virus infection.
Read more:
PA-X
Levene et al., Journal of Virology, 2021
Gaucherand, Porter et al. Cell Reports, 2019
Levene and Gaglia, Viruses, 2018 (review)
Rivas, Schmaling and Gaglia, Viruses, 2016 (review)
Khapersky, Schmaling et al. PLOS Pathogens, 2016
SOX
Gaglia et al. PLOS Pathogens, 2015
Gaglia et al. Journal of Virology, 2012
Covarrubias, Gaglia et al. PLOS Pathogens, 2011
xxx
Caspase-mediated regulation of interferon responses (in collaboration with Alexei Degterev)
Interferon responses are the key first line of defense against viruses. Surprisingly, reactivation of the oncogenic herpesvirus KSHV from latency does not elicit a strong type I IFN response. We have exciting new data that during infection, the host enzymes caspases are usurped by the virus to block interferon responses, another example of the virus turning core host pathways against itself. Caspases are druggable targets and could be exploited to potentiate innate immunity against this and other viruses to suppress viral replication. We are currently investigating how caspases control IFN and how KSHV controls caspases.
Read more:
Tabtieng et al., Journal of Virology, 2018
Tabtieng and Gaglia, Journal of Virology, 2018 (review)
xxx
Identification of novel regulators of gene expression encoded by Kaposi’s sarcoma-associated herpesvirus (KSHV)
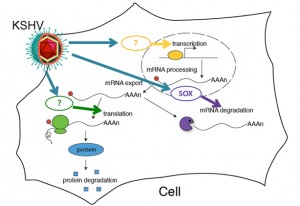 KSHV is a large DNA virus of the gamma-herpesvirus subfamily that cause Kaposi’s sarcoma and two rare types of lymphomas in individuals with weakened immune systems. It is one of the top causes of cancer death in Subsaharan Africa. In the US 1 in 200 transplant recipients develop Kaposi’s sarcoma.
KSHV is a large DNA virus of the gamma-herpesvirus subfamily that cause Kaposi’s sarcoma and two rare types of lymphomas in individuals with weakened immune systems. It is one of the top causes of cancer death in Subsaharan Africa. In the US 1 in 200 transplant recipients develop Kaposi’s sarcoma.
During replicative (lytic) KSHV infection, the transcriptome of the infected cells is severely altered. This is proposed to contribute to immune evasion and tumorigenesis, but the viral factors that govern these changes are poorly understood. Also, many of the ~90 open reading frames encoded by KSHV have uncharacterized functions. Using an overexpression screen, we have identified several previously uncharacterized KSHV proteins that may regulate host gene expression. These novel functions may be conserved among gamma-herpesviruses, as homologs of the candidate regulators in MHV68, a related mouse virus, also modulate gene expression. We are currently investigating how several of these putative regulators interact with cellular pathways, what genes they affect and what role they play in the viral life cycle using viral mutants.
Read more:
Butnaru and Gaglia, Curr Clin Microbiol Rep, 2019 (review)
xxx
Methods to analyze RNA-seq data
In collaboration with Christopher Rycroft at Harvard University, we are developing computational methods to analyze RNA-seq data, specifically data generated through 5′ degradome methodologies.
Read more:
Gaglia et al. PLOS Pathogens, 2015
(If you would like to read any of our papers and do not have access, please email us: Marta.Gaglia@tufts.edu)