Tiffani defended her thesis! Congratulations, Dr. Hui!
Yu-Shan Lin
Tiffani received the Award for Outstanding Academic Scholarship from the Graduate School of Arts and Sciences! Congratulations, Tiffani!
YSL was promoted to Professor. She also became the Chemistry Department Chair.
Our paper “Assessing the performance of peptide force fields for modeling the solution structural ensembles of cyclic peptides” is published in J. Phys. Chem. B.
Our paper “The immune-evasive Proline 283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure,” with the Shoulders Lab at MIT is published in Sci. Adv.
Tiffani has been selected as a DESRES Doctoral Fellow! Congratulations, Tiffani!
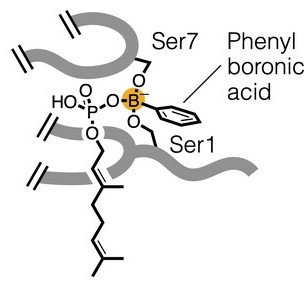
Our paper “A boron-dependent antibiotic derived from a calcium-dependent antibiotic” with the Chu Lab at the National Taiwan University is published in Angew. Chem.
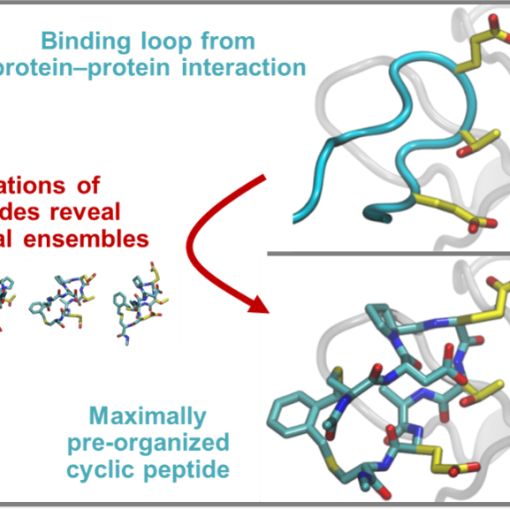
Our paper “Computational prediction of cyclic peptide structural ensembles and application to the design of Keap1 binders,” with the Kritzer Lab at Tufts is published in J. Chem. Inf. Model.
Our paper “Genetically-encoded discovery of perfluoroaryl-macrocycles that bind to albumin and exhibit extended circulation in-vivo,” with the Derda Lab at the University of Alberta is published in Nat. Commun.
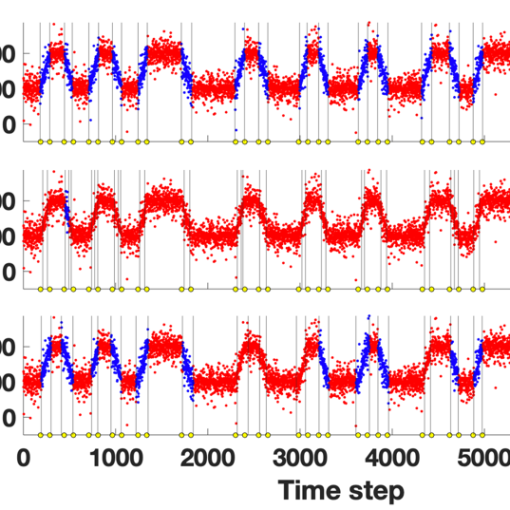
Our paper “Modeling changes in molecular dynamics time series as Wasserstein barycentric interpolations,” with the Murphy Lab at Tufts is published at the SampTA 2023 Conference.
Francini Fonseca Lopez defends her PhD thesis. Congratulations, Francini!
Marc Descoteaux completed his 4+1 program and graduated with a M.S. in Materials Science and Engineering. Congratulations, Marc!
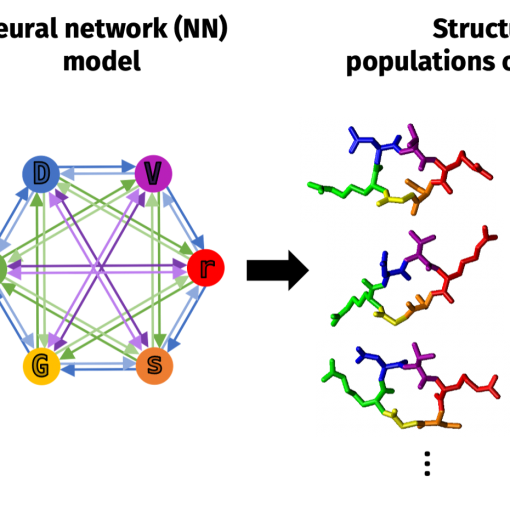
Our paper “Training neural network models using molecular dynamics simulation results to efficiently predict cyclic hexapeptide structural ensembles,” is published in J. Chem. Theory Comput.
Jovan Damjanovic defends his PhD thesis. Congratulations, Jovan!
The YSL lab presented at the Tufts Open House!
YSL receives the Tufts University Rising Innovator Award.
YSL receives the Annual Women Leadership Award from the Rotary Club of Cambridge.
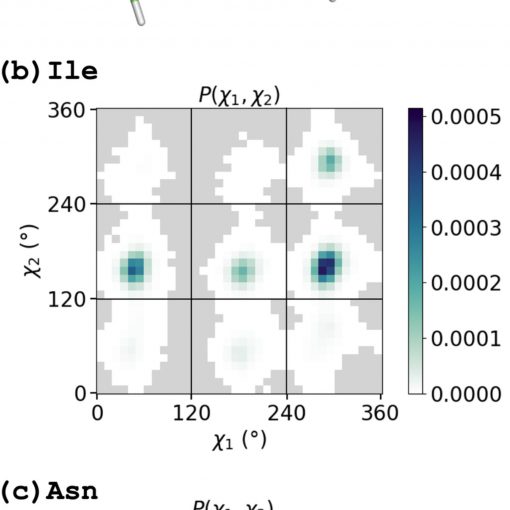
Our paper “A backbone-dependent rotamer library with high (φ, ψ) coverage using metadynamics simulations,” is published in Protein Sci.
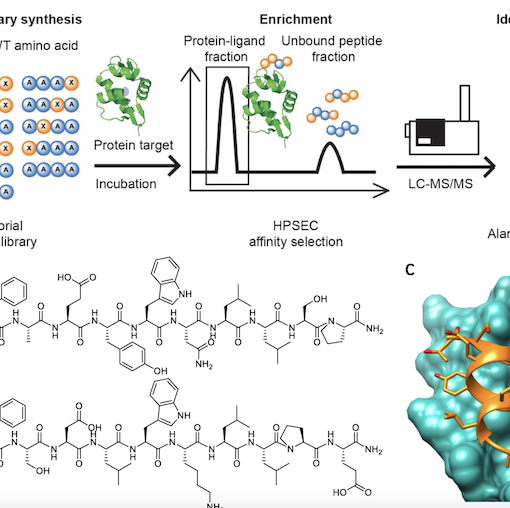
Our paper “Binary combinatorial scanning reveals potent poly-alanine-substituted inhibitors of protein—protein interactions” with the Pentelute Lab in the Department of Chemistry at MIT is published in Comm. Chem.
Marc Descoteaux wins the ACS undergraduate award in physical chemistry! Congratulations, Marc!