Mechanism of ALAD
δ-Aminolevulinic Acid Dehydratase (ALAD) is considered an aldolase, but it has a unique mechanism. ALAD appears to combine the two classes of aldolases in a complex mechanism, involving the formation of two Schiff bases, catalyzing an asymmetric condensation, and cyclization to form porphobilinogen (PBG).40
The substrates are two molecules of ALA, which are associated with an A-site and P-site (in regard to their place in the final product, A refers to acetyl and P to propionyl).45 The binding of A-site ALA prompts the active site “lid” to close, and this ALA likely coordinates with zinc.45
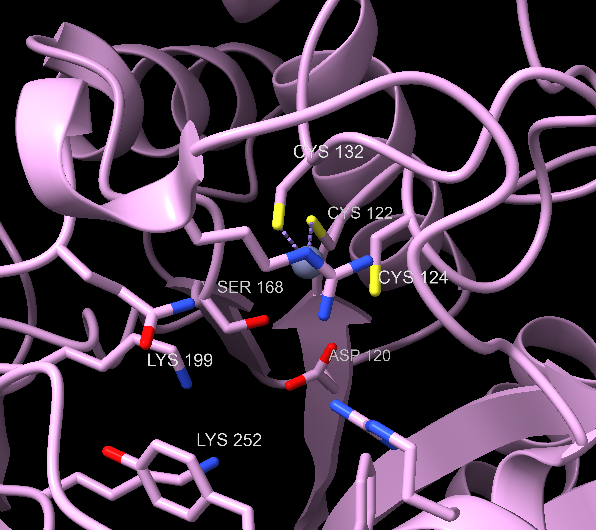
Two enzyme-substrate Schiff base intermediates are made. There is not yet a consensus on which one is formed first.50 The P-site ALA forms a Schiff base with Lysine 252, the A-site ALA with Lysine 199.53 There is also controversy whether the CA3-CP4 bond or CA4-NP bond forms first, along with the protonation states of the various Schiff base intermediates. For the purpose of this page, we present a mechanism that is widely favored, supported by recent physical chemistry analysis (as adapted from Ref. 45), and specific to the human ALAD active and metal binding site.45,53
Assuming the P-side ALA forms a Schiff base first (releasing a water molecule), the next step is the formation of the A-side ALA Schiff base intermediate with Lys199, from its zwitterionic form (Step 1). Then, the carboxyl of the A-side ALA abstracts its own pro-R proton, making an enamine (Step 2).
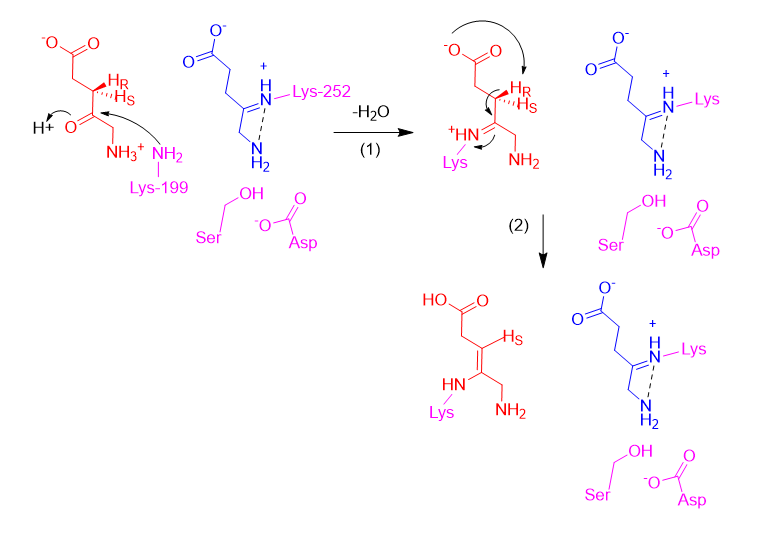
In the third step, the enamine acts as a nucleophile and attacks the Schiff base intermediate for the P-side ALA, creating a bond between CA4 and CP4. Then in Step 4, upon deprotonation by an active site serine (prompted by its deprotonation by aspartate), the P-side ALA amine attacks the A-side ALA Schiff base, making the CA4-NP bond. A five membered ring has formed, but it is still covalently bonded to the enzyme.
Next (Step 5), the amine of Lys199 deprotonates its own carboxyl. In Step 6, the amine of Lys199 is freed as an internal Schiff base is formed. Then (Step 7), the free amine abstracts the proton from CA3, creating the double bond between CA3 and CA4.
A similar electron-pushing sequence is likely repeated in Step 8-10 to remove Lys252 and form the double bond between CP4 and CP5, but the same proton abstraction from the carboxyl and Schiff base formation to free the Lys252 amine is not possible here due to its position. Since the mechanism from the Step 7 intermediate to PBG is not explicitly detailed (in Ref. 12), we have proposed that the amine of Lys252 abstracts the proton from the active site serine (which subsequently abstracts aspartic acid’s proton and restores the active site) (Step 8). Then, a general base abstracts the proton from CP5, forming a double bond between CP4-CP5 and freeing Lys252 (Step 9). While Ref. 45 shows that Lys252 becomes protonated, suggesting it may abstract this proton, we were unable to propose a logical mechanism for this. Potentially, the general base could later exchange the proton with Lys252 (Step 10).
In summary, ALAD functions as an aldolase, catalyzing the asymmetric addition of two ALA molecules into PBG using two Schiff base intermediates. The zinc is likely involved in substrate binding, but there is not recent experimental evidence to support a direct role in the catalytic mechanism. The displacement of this zinc likely precludes substrate coordination and Schiff base formation, thus halting the enzyme’s action. Thus, PBG synthesis and heme biosynthesis cannot proceed.
December 5, 2021 at 9:32 pm
Great job overall! You do mention specific amino acids of ALAD throughout the paper and a figure that describes or shows the enzyme could help visualize what the enzyme active site physically looks like. However, if you can’t find one, I don’t think it would take away from this section at all.
December 6, 2021 at 10:13 pm
This page does an excellent job of describing this mechanism. I like the comparisons to things we’ve talked about in class such as aldolases; it made it easier to understand exactly what’s happening throughout the mechanism. I also liked the descriptions of the different active sites and why they are referred to as the A and P sites, and I like the mention of the enzyme closing like a lid; makes it clear how the enzyme changes once the substrates are bound. Finally, I liked that this page was clear about what parts we are certain about in the mechanism and what parts are still under investigation; I found that interesting.
December 7, 2021 at 12:08 am
I agree with my group mates. I thought the explanation behind all of the mechanisms was very thoughtful and thorough. I don’t have much to add to this section!
July 22, 2025 at 5:36 am
Danke Digital olarak; web tasarım, sosyal medya reklamcılığı, Google Ads, SEO ve yapay zeka destekli yaratıcı çözümler gibi birçok alanda markalara özel çözümler üretiyoruz.