Three examples of affected proteins we have are Calmodulin, PKC, and Synaptotagmin. Calmodulin and PKC are both messenger proteins, while Synaptotagmin is a membrane trafficking protein. Lead binding interrupts all three protein’s signaling capabilities, which leads to defects in the nervous system.
Introduction of Lead to Calcium Binding Sites
Lead poses significant health threats because of its ability to mimic the effects of metal ions, such as zinc and calcium, that are used for biological signaling and metabolic processes. Focusing on how calcium is affected by lead, Pb2+ mimics and binds in protein active sites, displacing of calcium.2 Lead can mimic calcium because they share similar properties. In its +2 cationic form, lead has a radius of 132 pm while the calcium cation has a radius of 106 pm.3 In their elemental forms, lead has a radius of 175 pm and calcium has a radius of 197 pm.3 Protein active sites are able to accommodate lead instead of calcium due to their similar radii and shared +2 cationic form. Two highly affected proteins that modulate the secretion of calcium are calmodulin, a multifunctional intermediate calcium-binding messenger protein, and Protein Kinase Cα (PKC), a kinase involved in calcium-mediated signal transduction.5
Protein Kinase Cα (PKC) and Calmodulin
As seen in Figure 1, when lead binds to PKC it alters membrane-binding properties of C2α, a peripheral membrane-binding site of PKC.5 Normally, in response to Ca2+ binding to the PKC isoenzyme, the C2 domain is anchored to the parent enzyme in the lipid membrane.2 This binding is essential for kinase activation which controls signaling pathways essential for cell proliferation, differentiation, and survival.2 However, the Pb2+ binding inhibits the PKC activity. NMR experiments have determined that Pb2+ binds to C2α with higher affinity than Ca2+.2 Also, FRET spectroscopy results illustrate Pb2+ displaces Ca2+ from C2α through high-affinity interactions with the membrane-unbound C2α.2
Signaling proteins like PKC and calmodulin regulate cell activity by responding to altering levels of calcium.6 When lead takes calcium’s place in the calcium binding domains of these proteins, the proteins cannot react to calcium levels and therefore disrupt the electrostatic properties of the protein. Structurally for PKC and calmodulin, regardless of whether Ca2+ or Pb2+ bind to the active site the protein does undergo a very minor conformational change.7 Due to sensitivity of the amino acid residues in PKC, when Ca2+ or Pb2+ binds it caused a structural change by altering the N-H backbone or carboxyl side chain functioning as the metal site ligand.2 Although there is a minor conformational change, as seen in Figure 1 and 2, the crystal structures of lead and calcium for bound and unbound PKC and lead and calcium for bound and unbound calmodulin are practically superimposable. This indicates that the significant effect of lead binding is the electrostatic properties of the binding site on PKC.
Synaptotagmin I
In general, synaptotagmins are membrane-trafficking proteins, which means they control what goes in or comes out of a membrane.12 When calcium binds to a synaptotagmin, neurotransmitters are released. There are a variety of neurotransmitters released by synaptotagmins, so they are considered a family of different proteins.12 Synaptotagmin I is the primary synaptotagmin of interest because it signals the SNARE complex.13 The SNARE complex is a large protein family which mediates neurotransmitter release of synaptic vesicles.14 If lead binds instead of the intended calcium, it disrupts the synchrony of neurotransmission.14 This means that spontaneous neurotransmitter release is increased while evoked release is decreased.
Structure and Binding sites of Synaptotagmin I
Synaptotagmin I has an N-terminal segment that connects the protein to a synaptic vesicle and it is a transmembrane helical domain.13 It also has two cytosolic binding sites that are in the C2A and C2B domains.13 These domains bind to anionic phospholipids and are peripheral membranes.13
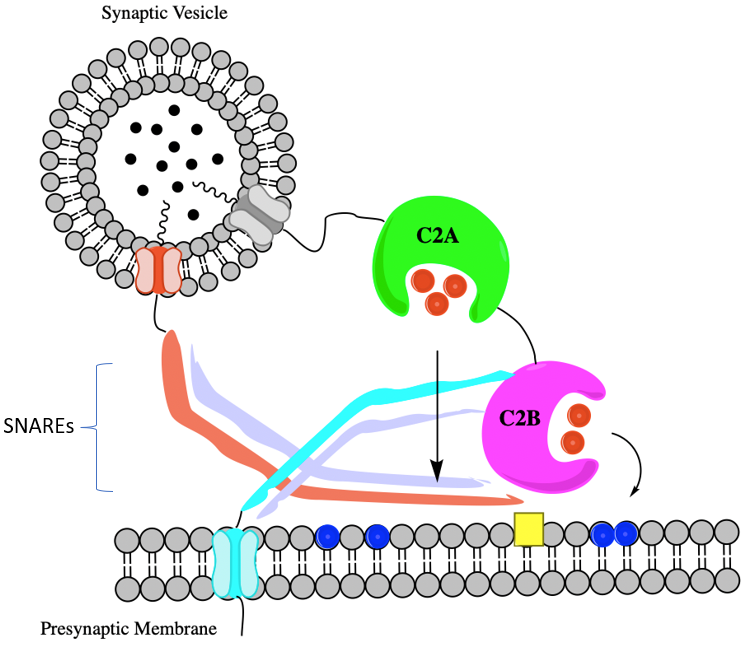
In the brain, lead binds to Synaptotagmin I.13 This protein has two domains C2A and C2B.13 The lead targets Ca2+ coordination sites that are oxygen-rich.13 Lead in both C2A and C2B bind to Site 1 which is a high-affinity lead binding site. There is a preference for lead to bind to in C2A over the C2B domain.13 This preference is because the C2A domain’s aspartic acid side chains are oriented towards the metal ion (in this case lead).13 Other conformational changes that exist between C2A and C2B are minimal and do not have an effect on binding preference.13
Lead Binding: Advantages and Effects
Lead has an increased affinity for both C2 domains in comparison to calcium, meaning lead will bind more tightly than calcium.13,14. As shown in the figure below, while lead and calcium have equivalent kon values, the koff value for lead is much smaller.13 This mixture of both fast binding from lead and then slow dissociation from the binding site creates a thermodynamic and kinetic trap that prevents calcium from binding.13 Lead binding to these domains is extremely exothermic, and there is favorable enthalpy so ΔG is a large negative value. This means this process occurs spontaneously.13 However, for calcium binding, the ΔG is largely impacted by entropy. The enthalpy does not have a large contribution because the binding of calcium is only slightly exothermic.13 In calcium binding experiments it was noted that there was no evidence of lead being displaced from Site 1 even when calcium was in excess.13

Regardless of the domain where lead binds, lead has an inhibitory effect on calcium binding, even if the other domain is completely vacant. As well, seen in Figure 5, in the domain itself, the lead binds to site 1 and while sites 2 and 3 are vacant calcium will not bind to these vacant sites due to lead’s inhibition. When bound to these domains, lead causes Synaptotagmin 1 to bind to phospholipids.13 In addition to directly competing with calcium and inhibiting calcium binding, lead causes the inhibition of Ca2+-dependent evoked release of neurotransmitters. Simultaneously, the presence of lead can increase the spontaneous neurotransmitter release.14 It is thought that lead alters synaptic activity due to blockage of the presynaptic calcium channels.14
I think the information presented here is good, but one thing that I’d change is the syntax of this section. Specifically, the paragraph of the lead binding location doesn’t have a lot of sentence length variety, which can some off as choppy (4 and 5 in particular of this section can be combined for example). Variety in sentence length throughout this paper could be improved but the information is pretty detailed so at the end of the day, this is just a personal preference and if you don’t want to change it I completely understand.
The reworking of a couple unclear sentences and the addition of some words/phrases to help sentences flow together (not only literal transition words like ‘then’ and ‘next’, but also words to communicate cause and effect) would go a long way to making this page more reader-friendly. The figures are helpful but maybe could use some labelling
Hi, Which figures are you referring too that need labeling or do you mean all of the figures?
For me personally, I felt like this section could’ve been more clear if you provided an initial cursory overview of all of the proteins that Pb2+ was affecting. My reasoning for this is that you initially talk about calmodulin and Protein Kinase Cα (PKC), but you then start talking about Synaptotagmin I. The transition from talking about the calmodulin and the kinase to talking about the Synaptotagmin I seems a little abrupt. Maybe an initial paragraph just explaining that lead can impact these three different proteins will make it easier for the reader to expect what’s coming later in the writing. Other than that, I felt that the writing was good!
I agree with Henry’s statements about reworking some unclear sentences and about labeling the figures. Figure 3 in particular could use some clearer labeling; I was confused on what was what in the figure and was having a hard time piecing it together even with the text referring to figure 3. However, the other figures are very well done. I liked how figures 1 and 2 had the lead-bound and calcium-bound proteins side-by-side so it was very easy to see the conformational differences and figure 6 provided a really nice summary of this section. Figure 4 also very clearly shows the aspartic acid residues, which goes along nicely with the description. I’m curious whether the permanence of lead binding to these proteins is related to them having a low enzyme turnover rate and how long the poisoning lasts after exposure has stopped.
Please, I want to know: is there a direct relationship between lead and acetylcholinesterase activity? i want to have more undestanding of it and I’ve searched for a while and the pdfs i come across are institution restricted.