Opioid Receptor Binding Assay
Background and Assay Principle
The β-arrestin GPCR assay measures the activation of G-protein coupled receptors using a luciferase reporter system (19). The luminescent product of luciferase activity, in this assay, is used to measure ligand binding to the GPCR as a way to identify potent κ agonists. The assay uses “fragment complementation technology” which uses an enzyme split into fragments (donor and acceptor), that will join and produce an active form of the enzyme when brought into close proximity. The activated enzyme can then be used to produce a measurable signal.
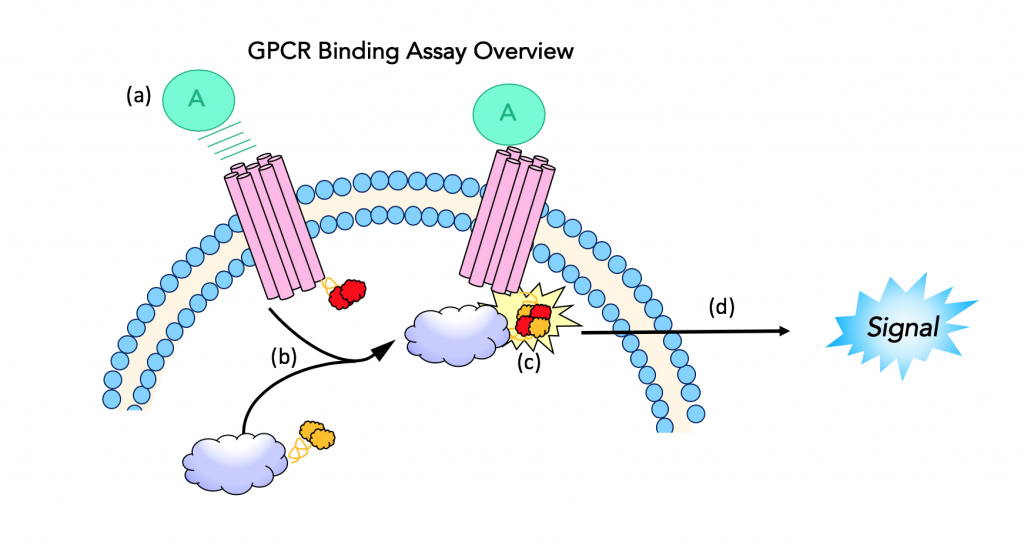
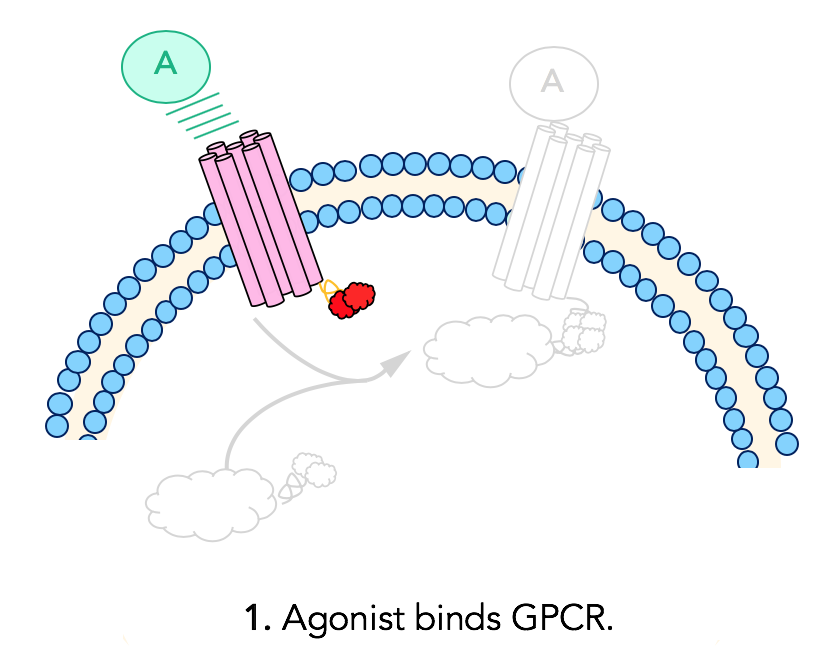
Step 1: Ligand binding
In this assay, complementary fragments of β-galactosidase are fused to the G-protein coupled receptor and β-arrestin, two important players in opioid signaling. The cell line used in this experiment expresses opioid receptors that are fused to a peptide fragment of β-galactosidase. These cells also express β-arrestin that is fused to the second, complementary enzyme acceptor fragment. The assay design begins with ligand binding to the GPCR. The ligand is the molecule of interest, which for this purpose is the synthetic opioid or the peptide opioid being studied.
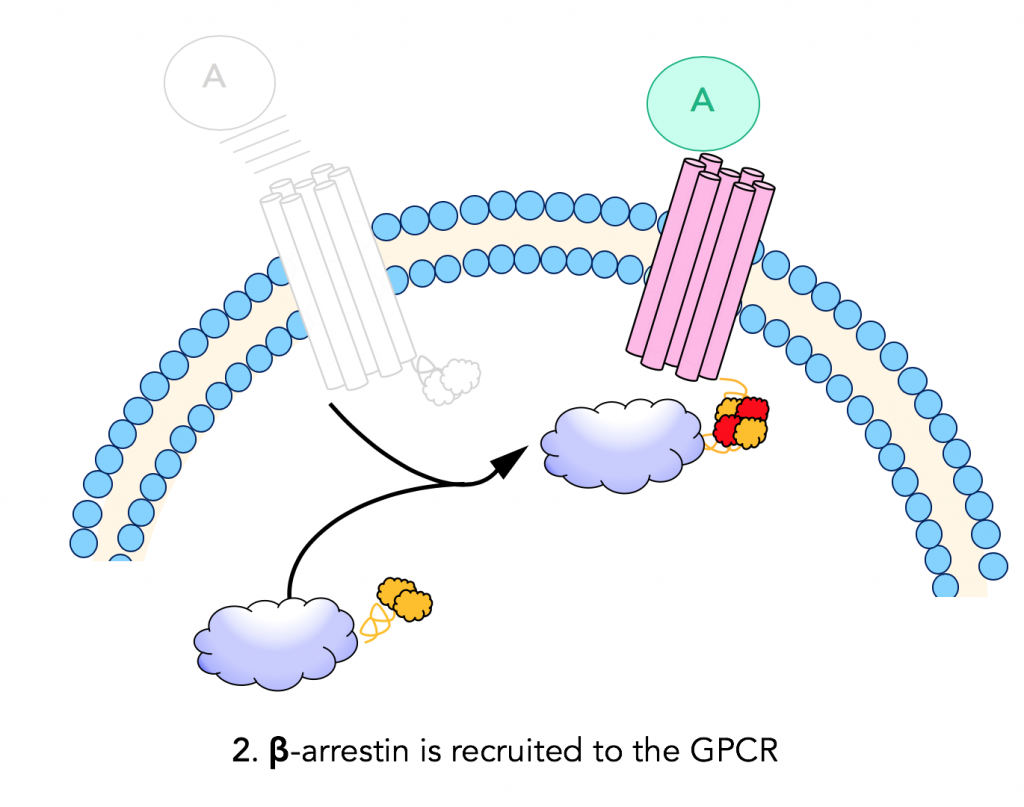
Step 2: β-arrestin recruitment
When a ligand binds and activates a G- protein coupled receptor, it recruits β-arrestin. β-arrestin is fused to the complementary enzyme acceptor (EA) fragment of β-galactosidase; and when it is recruited to the GPCR, the two enzyme fragments (enzyme donor and acceptor) are brought into close proximity.
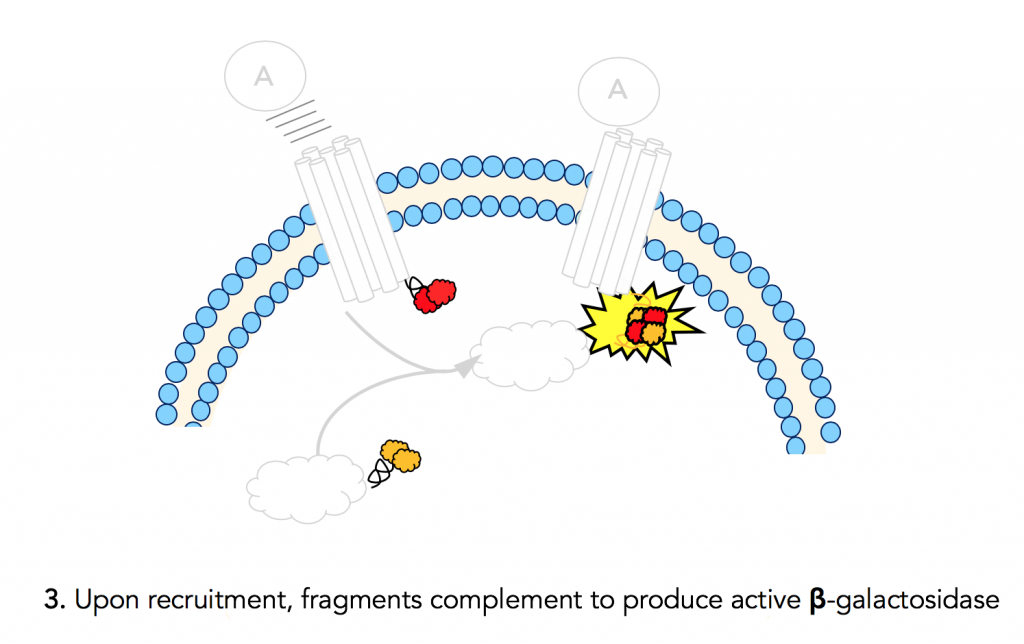
Step 3: Fragment complementation results in active β-gal
The ProLink™ peptide fragment of β-gal fused to the GPCR joins with its enzyme acceptor counterpart that is fused to β-arrestin. This results in an active β-galactosidase enzyme. The chemistry done by this activated enzyme is important for detection and quantification.
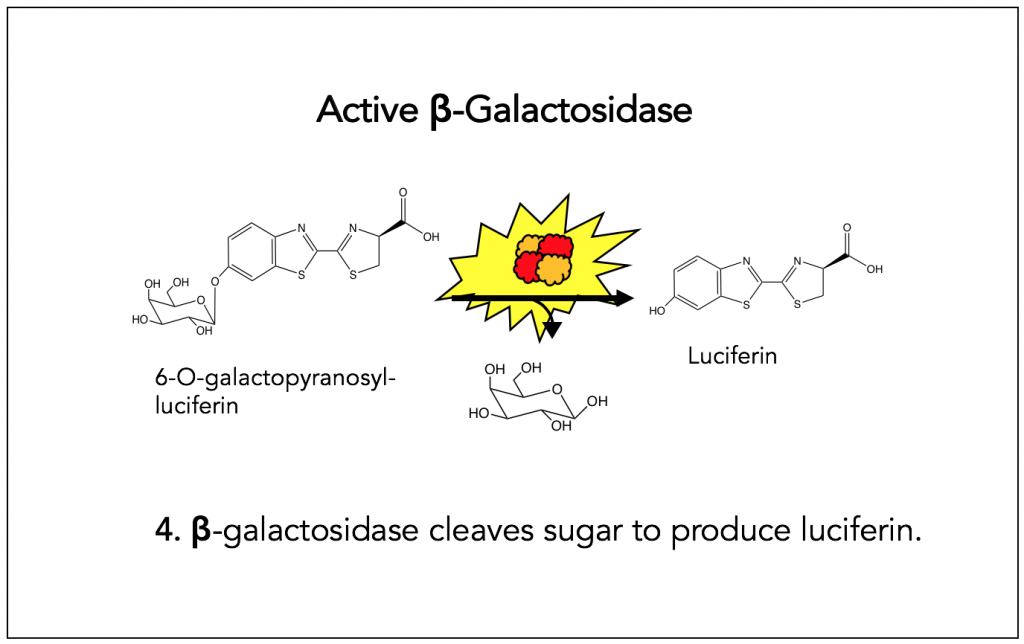
Step 4: β-Galactosidase cleaves sugar to produce luciferin
Fragment complementation of β-Galactosidase is important because of the chemistry performed by the active enzyme. More specifically, active β-galactosidase cleaves the sugar from 6-O-galactopyranosyl-luciferin to produce luciferin. The starting molecule in this reaction is added to cells as a part of the detecting agent.
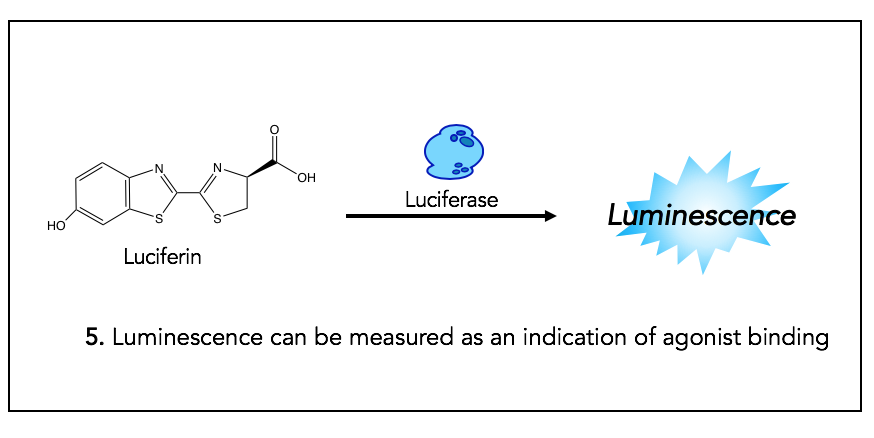
Step 5: Luciferase activity provides luminescent signal
The product of step 4, Luciferin, is the substrate for Luciferase, which is an enzyme used in many biochemical assays. The result of luciferase activity is a luminescent signal, which allows for quantification of the opioid agonist binding (given that the production of the substrate for this reaction is the result of GPCR activation).
Measurements and readout
The opioid receptor binding assay yields an EC50 value for the molecules tested. This value is termed the “half-maximal effective concentration” of the molecule of interest. In other words, the concentration of ligand at which half of the maximum signal is observed. A lower EC50 indicates better binding affinity. Typically, EC50 values are determined by fitting a dose response curve to the data.

Applications for opioid peptides
This assay has been used to quantify the efficacy of different peptide opioids being explored as less addictive alternatives for traditional pain management drugs. For example, Hughes et.al. used this assay to determine the most potent peptides in their library of κ agonists. Using this assay, they identified their most interesting derivative, JT09 for further investigation which had an EC50 of 29.9 nM(20, 21). We discuss this finding further under “Recent Research.”
November 9, 2019 at 11:14 pm
This page is excellent. I love how the images are after every small section and explain what you’re talking about. I would probably state that luciferase emits light when used first so that people know how this assay works. I was also a little unclear what this assay was testing for at first. A short summary of why this test was done in a sentence or two at the beginning would help me to know what I’m looking for
November 10, 2019 at 8:48 pm
I found this page to be extremely easy to follow. I really like the way the the images work with the paragraph to really solidify the knowledge of what is occurring. I do however agree with Paige that I did not know what the assay was looking for until the end. I would maybe explain why this test is needed and what the results would be looking for in the summary paragraph.