IKKβ and the Phosphorylation of IkBα
IKKβ structure
IkB kinase (IKK) is a protein complex that is involved in the control of NF-kB transcription factors. IKKβ is the kinase subunit in this complex that plays a heavy role in the canonical NF-kB pathway that we are examining. IKKβ phosphorylates NF-kB inhibitor, IkBα, causing the degradation of this inhibitor and the release of NF-kB into the nucleus (23).
IKKβ itself is a dimer, often described as shaped like a pair of scissors (23). As seen below, a region of each dimer extends like a “blade” and other regions comprise the “handle”.
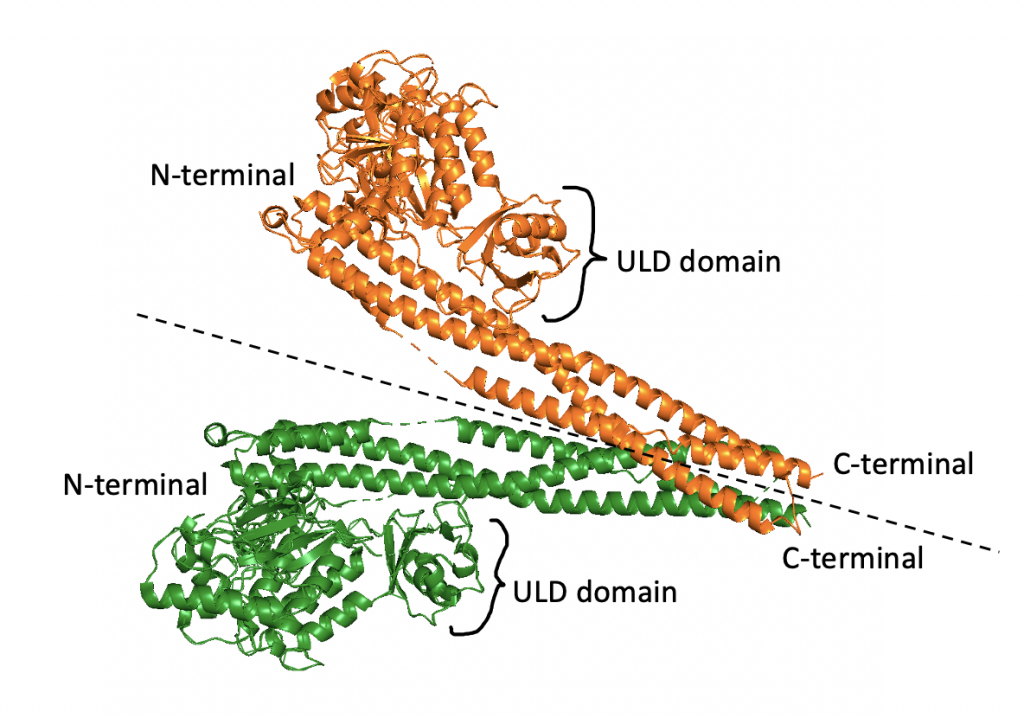
Both monomers of the IKKβ dimer have an N-terminal kinase domain (KD), ubiquitin-like domain (ULD), and C-terminal scaffold/dimerization domain (SDD). Each N-terminal is the kinase domain for the protein, meaning it is responsible for the phosphorylation activity of the protein. As suggested by the name, the SDDs, comprised of 6 alpha helices, are involved in the dimerization process. The ULDs contribute to the functionality and specificity of the dimer. In human IKKβ, the dimer is in an open configuration, where the SDDs are bent away from each other (23).
Within the dimer, only one of the KD regions is present in the active form. To explain this difference, Liu et. al. examined two characteristics of the KD region (23). Both KDs have an ATP binding pocket occupied by an inhibitor called K252a. This inhibitor is bound in an identical fashion in both domains (held in place by hydrogen bonds) (23). Therefore, this probably does not control KD activation.
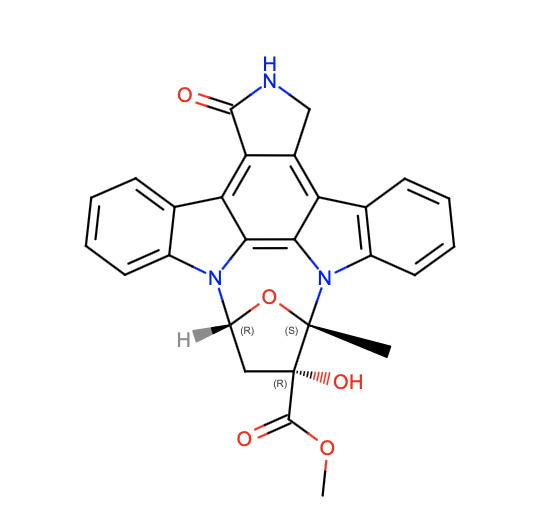
The other interesting characteristic is the phosphorylation of each KD. In this case, the dimer is asymmetric. One side has a phosphorylated KD, while the other KD will not be phosphorylated. Specifically, this involves the phosphorylation of Serine-177 and Serine-181 (with Serine-181 being the more critical residue for KD activity) (23). Phosphorylation status is possibly controlled by an upstream kinase or the stereochemistry of the dimer (23). Once phosphorylated, nearby residues help to stabilize the phosphate oxygen atoms and form the active site conformation. Without phosphorylation, the conformation of the active site does not allow for substrate binding. Therefore, Liu et. al. concluded that phosphorylation is the main factor involved in activating the KD (23).
IKKβ dimerization
Between the three domains in each monomer, Van der Waals interactions and hydrogen bonding between all three domains are responsible for stabilizing the structure (23). These monomers are then paired to create the dimer seen above. This interaction occurs between the SDD regions, namely at the α2, α3, and α6 helices of each monomer. The majority of the interface surface is comprised of hydrophobic residues (such as Phe and Leu) (23). This makes sense because hydrophobic residues tend to congregate in order to avoid water. However, there are also several hydrophilic residues within the region that comprises the interface (including Ser, Asp, and Gln). While this seems unfavorable, the energy is compensated by the hydrogen bonds that are then able to form (23). Each half of the dimer contributes different residues for a total of 10 hydrogen bonds between the dimers (23).
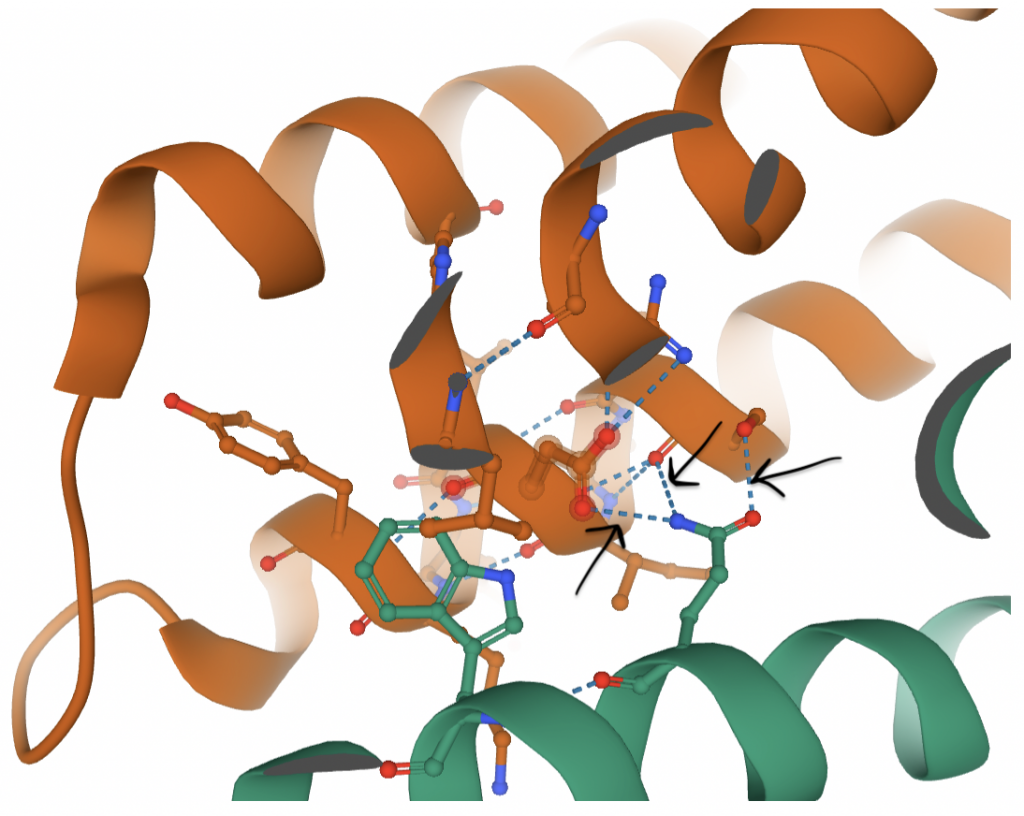
In the figure above, black arrows mark hydrogen bonds that contribute to the dimerization. Hydrogen bonds extend from atoms on the green residue (Gln-657) to atoms on orange residues. There are a handful of similar hydrogen bonds that all work together (in addition to hydrophobic interactions, not shown) to maintain the dimer.
Wu et. al. found in their research that dimerization is necessary for IKKβ activation, but once this occurs and the KD is phosphorylated, IKKβ can function normally as a monomer to phosphorylate IkBα (24).
Phosphorylation of IkBα by IKKβ
Activated IKKβ is able to fit IkBα into its active site. An area within the C-terminal region of IkBα, called the PEST region, assists with this binding process, ensuring that the orientation of the substrate within the active site is optimized. Once in the active site, the N terminal region of IkBα is phosphorylated at Serine-32 and Serine-36 (24).
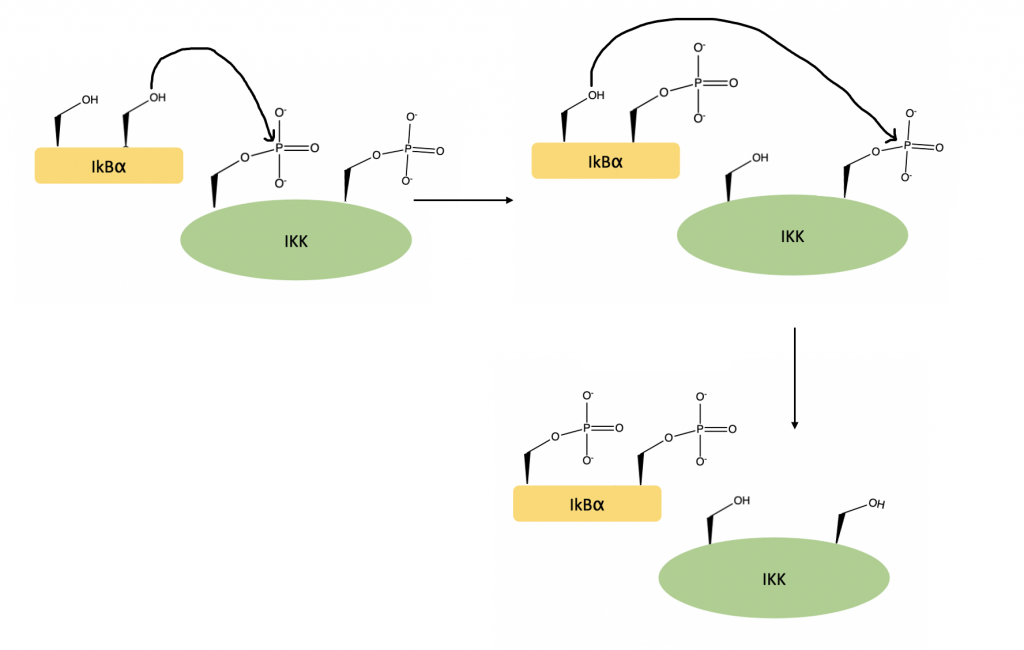
Once phosphorylated, a ubiquitin ligase complex targets IkBα for degradation. This frees NF-kB, which can move to the nucleus to activate gene transcription (25).
Recent Comments