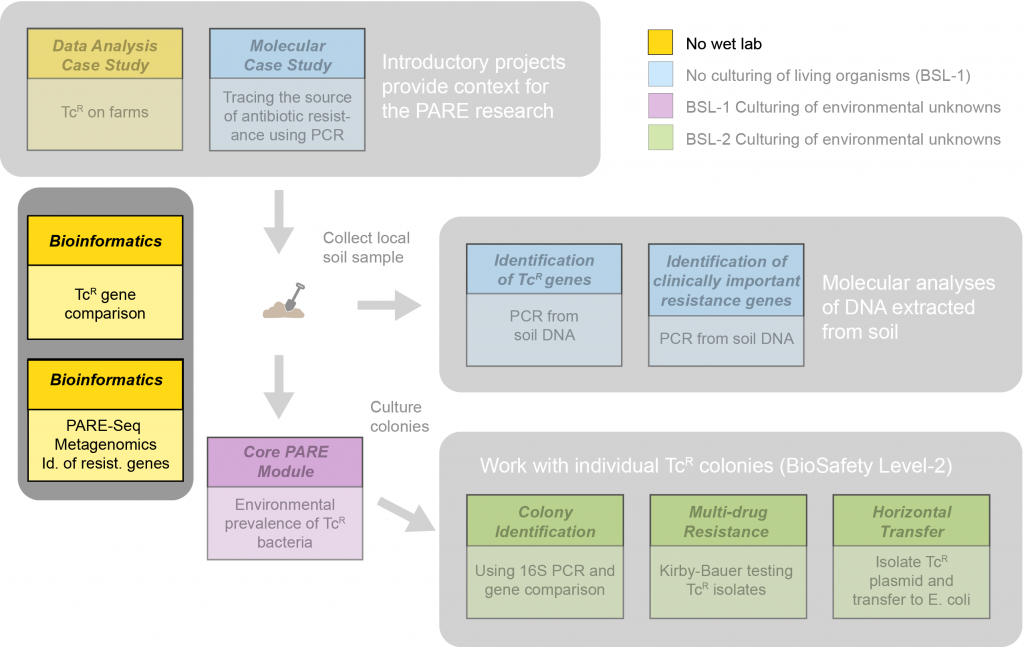
Metagenomic bioinformatics module: PARE-Seq
The metagenomic bioinformatics module, PARE-Seq, demonstrates how scientists can use total metagenomic DNA extracted from environmental samples to search for any antibiotic resistance genes that may be present, regardless of what organism carries them, and regardless of whether that organism is living or expressing (using) those genes. Unlike traditional bioinformatics teaching activities, a computer cluster is not required. We use the free online platform, Galaxy. PARE-Seq is a comprehensive suite of teaching material conceived and created by Scarlet Bliss, an undergraduate at Tufts University and in collaboration with Dr. Amy Pickering (Berkeley), Dr. Erica Fuhrmeister (Tufts University), and Eve Abraha.
Introduction to bioinformatics and sequence analysis using tetR genes
The Tetracycline Gene Comparison Module introduces students to the Basic Local Alignment Search Tool (BLAST) to compare DNA and protein sequences of tetracycline resistance genes.
In this computer-based activity, students will use web-based bioinformatics tools to perform DNA and protein comparisons of tetracycline resistance genes and their products.
This is intended to be a stand-alone case study. You do not need to complete other modules before starting this one, and it doesn’t have a wet lab.
This module was developed by Dr. Jennifer Larson, Capital University.
