To build important molecular skills, we offer introductory and advanced modules where students can learn micropipetting, PCR and DNA extraction skills. These modules also avoid working with resistant unknown organisms, since work is done directly from soil without culturing. Videos in the Background section provide useful context for students.
Molecular Case Study
Tracing the course of antibiotic resistance using PCR

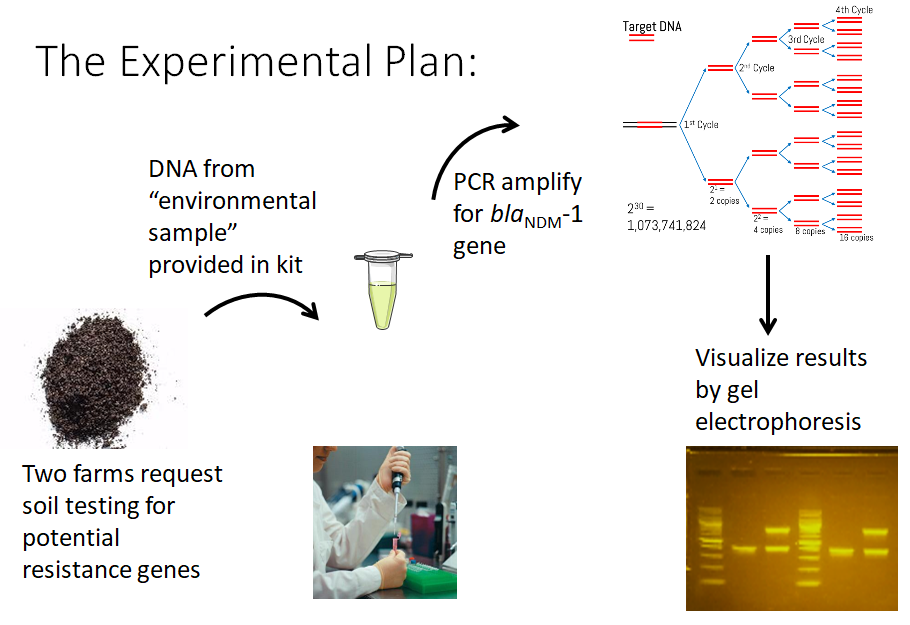
This skill-building lab represents a fictional case study of a very real problem. Students are presented with information regarding an outbreak of food-borne infection that is resistant to treatment with antibiotics. They are tasked with using PCR and gel electrophoresis to establish whether farms are at risk from antibiotic resistant bacteria that may be spreading in the environment. This known-outcome activity introduces students to the concept of environmental antibiotic resistance and is a great skill-builder to precede the “Identification of TetR Genes” (eDNA) molecular module.
Downloads
The most recent materials for this module (including instructor and student guide as well as teaching slides) can be found on the Curriculum Downloads page of the miniPCR Agricultural Monitoring Lab web page.
External Links
This module is a collaborative effort with miniPCR and is appropriate for high school and undergraduate classrooms.
Identification of TcR Genes
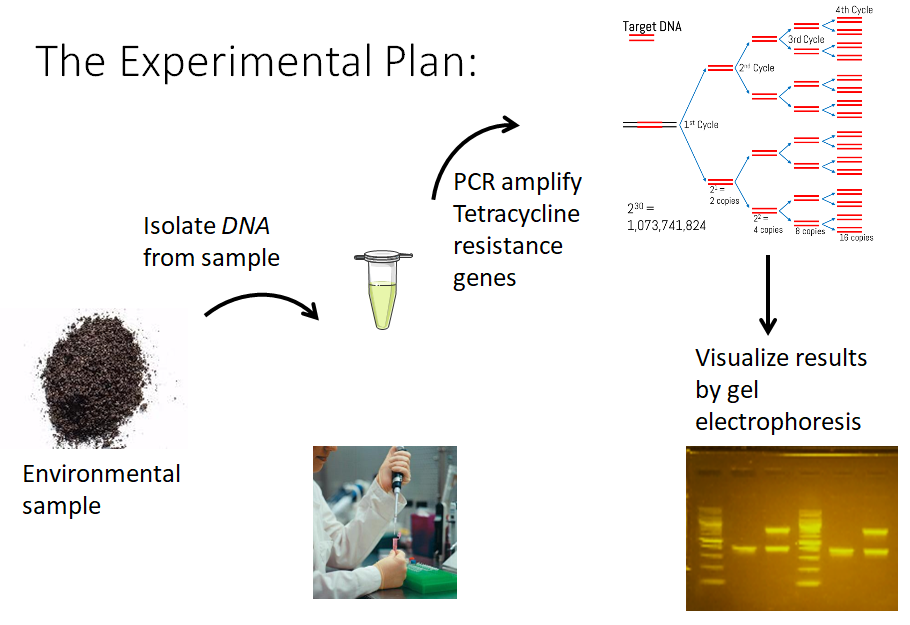
Identification of tetracycline-resistance genes from total soil DNA
This molecular-based approach to environmental surveillance allows students to accrue authentic data added to our database without the risks and complications of culturing environmental unknowns that are potentially multi-drug resistant. Students collect a soil sample and then, in the laboratory classroom, they extract total metagenomic DNA from soil samples and use PCR to test for presence of two different tetracycline resistance genes. Students upload soil site location information on their phones in the field at the collection site and then add more data about the resistance profile once they obtain PCR results.
This module was created in collaboration with miniPCR and with assistance from PARE instructors Dr. Adam Kleinschmit (University of Dubuque) and Dr. Jennifer Larson (Capital University).
Module Organization
This module doesn’t require handling of biological organisms; however, the same methods can be applied to TcR colonies isolated from the core PARE module, which would require BSL-2 handling of environmental unknown organisms.
Downloads
Data Upload Instructional Slides
The most recent materials for this module (including instructor and student guide, student worksheet and teaching slides) can be found on the of the Curriculum Downloads page of the miniPCR eDNA project web page.
Clinical Resistance in Soil DNA
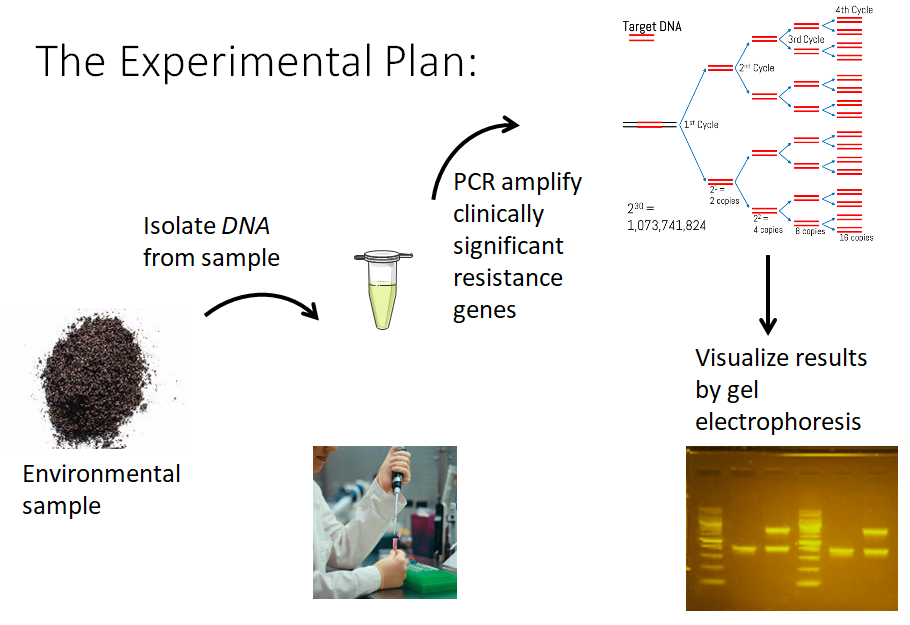
Identification of clinically important resistance genes
This module is currently in development. Students will use primers to detect genes representing antibiotic resistance types that are difficult to treat and recently emerging globally in patients with antibiotic resistant infections (e.g. blaNDM-1, mcr-1). The assumption is that these “emergent” genes are circulating in the environment before they are detected in patients. With enough student crowdsourcing, we believe we can identify geographic areas harboring high levels of these resistance genes which could then lead to localized stewardship efforts to contain or decrease the presence of the particular antibiotic resistance determinant.
The background video “Spotlight on Antibiotic Resistance Genes of Clinical Significance” is a good primer for this work.
Module Organization
This module is closely related to the “Identification of TcR genes” module; however the presence of genes targeted is expected (hoped) to be extremely rare. The curriculum for this module will be crafted in collaboration with experts in the Tufts Center for Integrated Management of Antimicrobial Resistance to ensure that messaging regarding identification of clinically important resistance markers is appropriate.
Acknowledgements
Development of this module is a collaborative effort with the miniPCR company, Dr. Adam Kleinschmit (University of Debuque), Dr. Jennifer Larson (Capital University), Dr. Amy Pickering (Tufts University, Engineering), and Dr. James Kirby (Harvard Medical School).