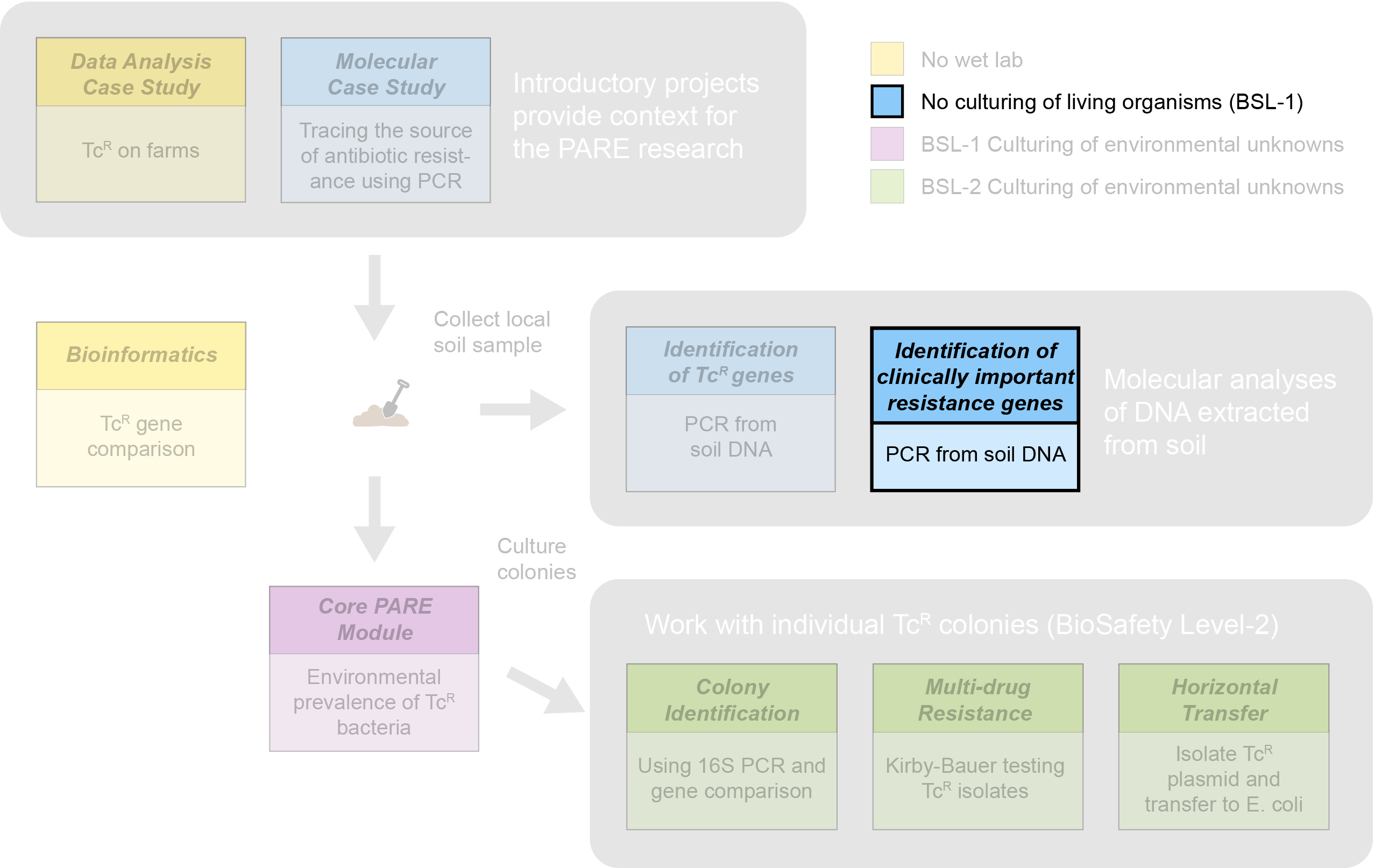
Identification of clinically important resistance genes
This module is currently in development. Students will use primers to detect genes representing antibiotic resistance types that are difficult to treat and recently emerging globally in patients with antibiotic resistant infections (e.g. blaNDM-1, mcr-1). The assumption is that these “emergent” genes are circulating in the environment before they are detected in patients. With enough student crowdsourcing, we believe we can identify geographic areas harboring high levels of these resistance genes which could then lead to localized stewardship efforts to contain or decrease the presence of the particular antibiotic resistance determinant.
Module Organization
This module is closely related to the “Identification of TcR genes” module; however the presence of genes targeted is expected (hoped) to be extremely rare. The curriculum for this module will be crafted in collaboration with experts in the Tufts Center for Integrated Management of Antimicrobial Resistance to ensure that messaging regarding identification of clinically important resistance markers is appropriate.
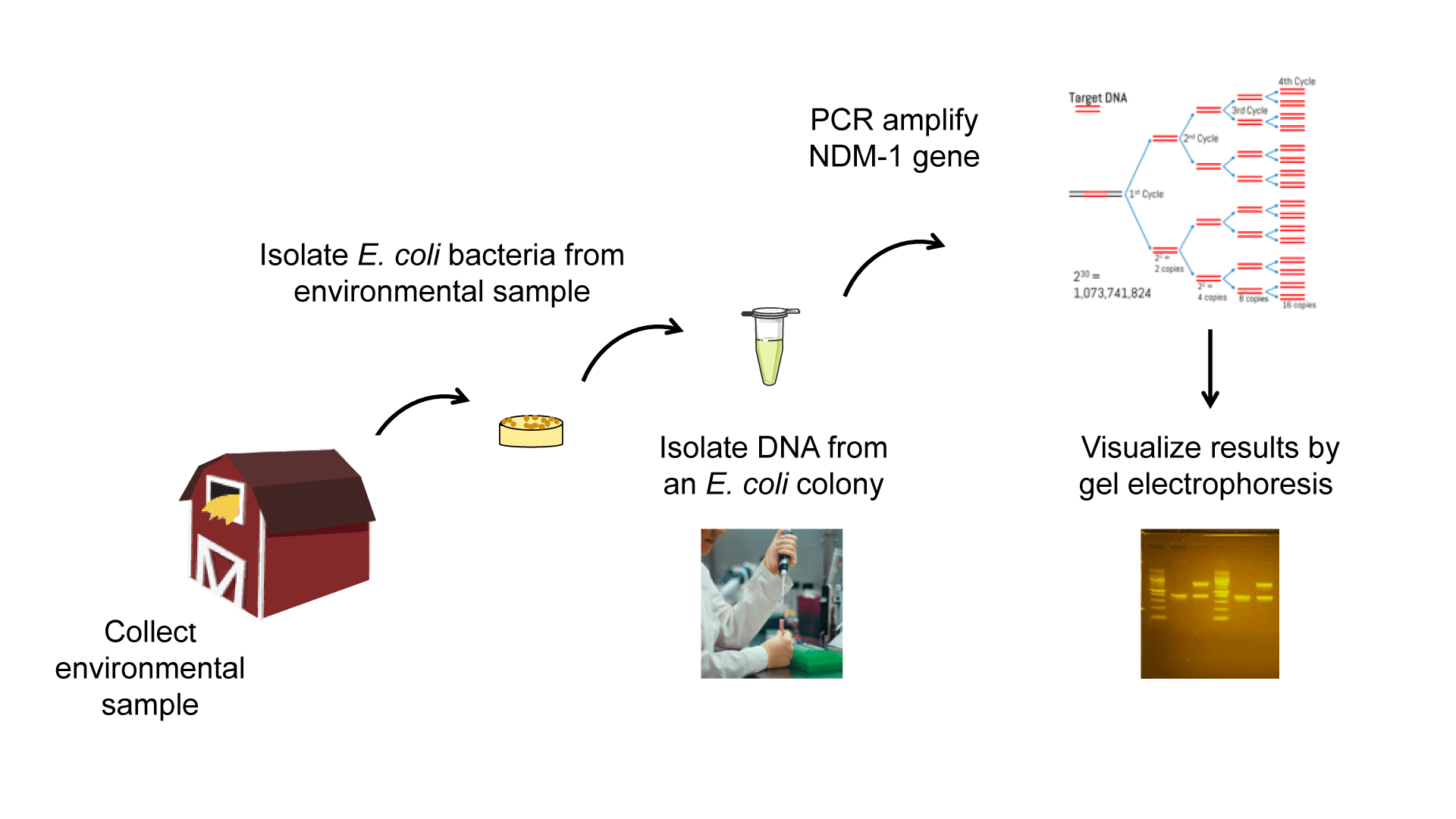
The Experimental Plan
Acknowledgements
Development of this module is a collaborative effort with the miniPCR company, Dr. Adam Kleinschmit (University of Debuque), Dr. Jennifer Larson (Capital University), Dr. Amy Pickering (Tufts University, Engineering), and Dr. James Kirby (Harvard Medical School).